Cytokinins (CKs) are a chemically diverse class of plant growth regulators, exhibiting wide-ranging actions on plant growth and development, hence their exploitation in agriculture for crop improvement and management. Their coordinated regulatory effects and cross-talk interactions with other phytohormones and signaling networks are highly sophisticated, eliciting and controlling varied biological processes at the cellular to organismal levels. In this review, we briefly introduce the mode of action and general molecular biological effects of naturally occurring CKs before highlighting the great variability in the response of fruit crops to CK-based innovations. We present a comprehensive compilation of research linked to the application of CKs in non-model crop species in different phases of fruit production and management. By doing so, it is clear that the effects of CKs on fruit set, development, maturation, and ripening are not necessarily generic, even for cultivars within the same species, illustrating the magnitude of yet unknown intricate biochemical and genetic mechanisms regulating these processes in different fruit crops. Current approaches using genomic-to-metabolomic analysis are providing new insights into the in planta mechanisms of CKs, pinpointing the underlying CK-derived actions that may serve as potential targets for improving crop-specific traits and the development of new solutions for the preharvest and postharvest management of fruit crops. Where information is available, CK molecular biology is discussed in the context of its present and future implications in the applications of CKs to fruits of horticultural significance.
- abiotic stress
- biotechnology
- food security
- micropropagation
- shoot proliferation
- somatic embryogenesis
- phytohormones
- plant growth regulators
- postharvest
- quality attributes
1. Introduction
Cytokinins (CKs) are a unique class of plant growth regulators (PGRs) with a long and interesting history. Their existence as compounds capable of inducing cell division in cultured plant tissues was first documented more than 100 years ago [1]. With the discovery of an increasing number of compounds with CK-like actions in plants even to date, CKs are thus broadly grouped as natural (purine-based molecules, which are either isoprenoid or aromatic CKs) or synthetic CKs, which are urea-based [2]. Figure1 shows the structural configurations of some existing natural and synthetic CKs. These CKs are considered to possess potential influence throughout the entire course of a plant’s life from embryogenesis until death in both lower and higher plants, as evidenced in the diverse physiological and biochemical functions during the life cycle of the different organisms [2][3][4]. They are involved directly or indirectly in different plant physiological processes such as the regulation of seed germination, shoot elongation and proliferation, induction of flowering, fruiting and seed set, and senescence [5][6][7][8][9][10]. Particularly, their roles in fruit set, delay of senescence processes—including fruit ripening and defoliation [11], which are concomitant with the release of buds from apical dominance [12], remain fundamental to the successful production of many horticultural fruit crops. Coupled with the development of genetically improved crop varieties and the application of improved agronomic practices, the use of PGRs including CKs has contributed positively to the green revolution and subsequent increase in agricultural productivity globally [13]. However, fundamental knowledge of the diverse roles of CKs in plants remains fragmented, and there is greater scope to deepen our knowledge of how CKs function and regulate cellular mechanisms that control plant growth and development. This knowledge will enable greater exploitation and application of CKs in horticultural fruit production. Recently, Koprna et al. [14] highlighted the potential of CKs as agrochemicals in pot and field experiments as they improve the growth dynamics and yields of a wide range of plants, including horticultural fruit crops
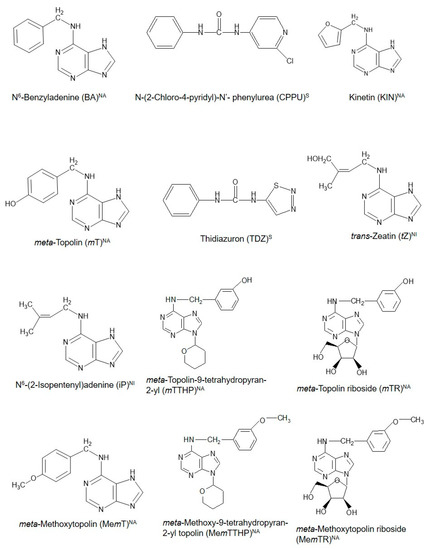
. Chemical structures of cytokinins (CKs) used in propagation, preharvest, and postharvest stages during the production of some common horticultural fruit crops. NA = natural aromatic CK; NI = natural isoprenoid CK, and S = synthetic CK
With more than 80 commonly known species of horticultural fruit crops available, their relevance to offset food and nutrition security concerns among the ever-increasing global population cannot be overemphasized. For centuries, horticultural fruit crops have been cultivated (mainly via conventional methods) as important dietary foods serving as the major sources of vitamins, antioxidants, and fibers for human needs [15][16]. As an indication of their economic and commercial values, the global production of the major fruit crops, including banana, apple, orange and mango, has witnessed a consistent and dramatic increase according to the Food and Agriculture Organization (FAO). Statistics from the FAO show that between 2000 and 2017, the production of mangoes, mangosteens, and guavas rose from 20 to over 40 million tonnes while banana production experienced a compound annual growth rate of 3.2% over the same period (http://www.fao.org/statistics/en/). Figures for banana cultivation were on a record high in 2017, reaching 114 million tonnes from 67 million tonnes in 2000. Other fruit crops such as oranges, apples, and grapes also showed positive trajectories in terms of their production, even though their incremental trends did not surpass that of banana (http://www.fao.org/statistics/en/). While these increases remain laudable, more effort to stimulate higher yield potential and the stability of the major fruit crops are needed to feed a world population that is predicted to reach 8 billion by 2025 [13]
The propagation of many fruit crops has intrinsic challenges such as low germination rate, heterozygosity of seeds, and prolonged juvenile phase, which hamper efficient and rapid growth [17]. Together with the changing climate, biotic and abiotic stresses can significantly influence productivity in major fruit crops [18][19][20][21][22][23]. In recent times, different strategies including genetic modification [19][24][25][26], encapsulation technology [20], photo-biotechnology [27], and the manipulation of phytohormone balances with compounds such as nitric oxide (NO) [28] and 1-methylcyclopropene (1-MCP) [29] have been explored to mitigate biotic and abiotic stresses. Furthermore, the systematic application of biostimulants, particularly plant growth-promoting rhizobacteria (PGPR) and mycorrhizal fungi have been demonstrated to hold the potential to mitigate biotic and abiotic stresses as well as boost fruit crop production [18][30]. In addition to these approaches, the diverse roles of PGRs, especially CKs, offer a potential avenue that requires more detailed attention [31][32][33][34][35][36]. As an example, Zalabák et al. [22] postulated that the genetic engineering of CK metabolism may offer greater potential to improve the agricultural traits of crops. In response to environmental cues, physiological and genome-wide microarray studies indicate an existing relationship with CK levels in planta [32]. In addition to increasing evidence of CKs’ influence in alleviating biotic and abiotic stresses, CKs play an important role in horticultural crop production where their application influences the morphological structure and nutrient content, as well as facilitates harvesting and the overall yield in a number of fruit crops [14][37]. Thus, in this review, we highlight and critically explore the potential of CKs in the propagation, growth, and general physiology with specific reference to some fruit crops. In the past three decades, the advent of molecular biology, genetic engineering, and exploitation of mutant technologies in various model plant species has led to a better understanding of the molecular mechanisms of CKs. Major breakthroughs in the 1990s led to the discovery of the CK signaling circuit networks that partly explain the diverse roles of CKs throughout a plant’s life cycle in molecular, cellular, and developmental contexts. Some of this research, mainly conducted in the non-horticultural
s species
a) as a model, has been comprehensively reviewed in the works of
- (a)
-
(a) Kakimoto [38], describing the perception and signal transduction mechanisms involving CK receptors in plants based on the molecular work conducted in the 1990s;
-
Kakimoto [38], describing the perception and signal transduction mechanisms involving CK receptors in plants based on the molecular work conducted in the 1990s;
-
(b) Hwang et al. [39], where CK–auxin relationships controlling early embryogenesis and organ differentiation and development are explained. The authors highlighted studies conducted in Lotus japonicus and Medicago truncatula that have led to the acknowledgement of the importance of CKs in nodule formation. Furthermore, the impacts of CK circuits in biotic and abiotic stress responses and regulation of senescence by CKs were critically described;
- (b)
-
(c) Steklov et al. [40], who compared the structural configuration of CK receptors and their phylogenetic relatedness across species including horticultural crops such as orange, apple, tomato, and grape.
-
Hwang et al. [39], where CK–auxin relationships controlling early embryogenesis and organ differentiation and development are explained. The authors highlighted studies conducted in Lotus japonicus and Medicago truncatula that have led to the acknowledgement of the importance of CKs in nodule formation. Furthermore, the impacts of CK circuits in biotic and abiotic stress responses and regulation of senescence by CKs were critically described;
- (c)
2. Metabolic Regulation of Cytokinin Activity
The metabolic production and control (biosynthesis, inter-conversions, and degradation) of CK homeostasis involve a wide range of enzymes [12][45]. Particularly, isopentenyltransferase
T) is an important enzyme involved in the first and rate-limiting step in CK biosynthesis that entails the transfer of an isoprenoid moiety to the
6 position of the adenine nucleotide [22][45]. An additional enzyme involved in the modification of CKs at the adenine part of the molecule was discovered in 2007. Evidence from the study by Kurakawa et al. [46] revealed the existence of a specific phosphoribohydrolase (designated as Lonely Guy;
G) in rice. The
G enzyme is responsible for the cleavage of ribose 5’-monophosphate from the CK nucleotides to form biologically active CK-free bases in one enzymatic step [22]. On the other hand, cytokinin oxidase/dehydrogenase
X) is central to the catabolism of CKs, where an irreversible cleavage of the CKs occurs, and the presence of auxins positively regulates this enzyme. Cytokinin oxidase/dehydrogenase is under a positive auxin regulation, leading to the regulated synthesis of CKs in plants and associated responses. The CK biosynthetic genes belong to a gene family that is developmentally and spatially regulated in its expression in plant cells [12][22]
Glucosyltransferases and xylosyl transferases catalyze
O-glucosylation,
N-glucosylation, and
O-xylosylation events, leading to the production of various CK conjugates whose full function remains to be completely characterized [47][48]. For instance, a recent evidence revealed the metabolic reactivation of
s-zeatin
tZ)
N-glucosides
7 and
9 positions) in
a, which is contrary to the previously-held hypothesis that
N-glucosylation irreversibly inactivates CKs [45]. Many of these enzymes involved in CK metabolism were discovered mainly in the 1990s through to the 2000s. The uridine diphosphate glycosyltransferases (UGTs) are now known to deactivate CKs such that the regulation of CKs in plants is precise during distinct developmental phases and in response to environmental conditions throughout the plant’s life [49]. Environmental factors, both abiotic and biotic, as well as endogenous inputs, tightly regulate the synthesis and degradation of CKs, generally, in plants [50]
3. Molecular Aspect of Cytokinin Actions
Molecular genetic approaches have been useful in unravelling the major sensing and signaling roles linked to CKs [12]. The CK receptors are of a histidine kinase (HK) nature with autophosphorylation events being important as part of the signaling transduction pathways that ultimately lead to the negative and positive induction of CK-controlled gene expression [39]. The CK signal pathway in plants uses a basic phosphorelay two-component system (firstly described in bacteria) which revolves around four sequential phosphorylation steps that alternate between histidine and aspartate residues, where a conserved CK-binding domain, Cyclases/Histidine kinases Associated Sensory Extracellular (CHASE), has an extracytosolic location [39]. The HK receptors are localized on endoplasmic reticulum (ER) membrane, and the CHASE domain lies in the direction of the ER, leading to the hypothesis that the in planta binding of CKs is in the lumen of the ER [47]. These CK receptors are part of a large family of transmembrane HK sensors with three main evolutionary branches in plants, which is evident through the application of various bioinformatics tools [40]. The cytokinin response factor
F) gene families are known to control cotyledon and leaf development. Although the
F genes, belonging to the family of AP2/ERF transcription factors, were first characterized in
s, they are found in all land plants [51]. The tomato-specific CRFs, termed
F genes, responds to CKs by controlling the development of leaf primordial and root tips, and they occur as two distinct clades [52]. The review by Cortleven et al. [53] highlights the importance of CK mutants in uncovering the signaling mechanisms and biosynthesis steps involved in the in planta production of natural CKs. For example,
G enzymes catalyse the reaction steps that increase the metabolic pool of CKs such as isopentenyladenine (iP) and
tZ in plant tissues [10][12]. The nuclear-localized type B response regulators (RRB or type B ARR) are transcription factors of the CK signaling pathway that CK targeted for gene expression [12][53]. Through a negative feedback loop, the other regulators, type A RRs (RRA), indirectly control the induction of the CK-responsive genes that are in fact targets of the type B RRBs [53]
As a result of the benefits associated with transgenic or genome engineering, desired traits can be manipulated in different horticultural fruit crops (Table 1), and this has largely been spurred on by accumulating new information on the molecular biological effects of CKs in plants. For instance, genes related to specific CKs such as CPPU
N-(2-chloro-4-pyridyl)
N´-phenylurea) and BA
6-benzyladenine) were recently identified in horticultural fruits. Following the treatment of pear fruitlet with 30 mg/L CPPU, the
R genes potentially influenced fruit development, bud dormancy, and light/hormone-induced anthocyanin accumulation [54]. The study by Ni et al. [54] indicated that CKs have the potential to stimulate the accumulation of anthocyanin in pear. Similarly, the upregulated expression of the
X gene contributed to the induction of anthocyanin content in strawberry treated with varying concentrations of CPPU [55]. Apart from the impact of CKs on specialized (secondary) metabolites [54][55], central (primary) metabolites, especially the carbohydrate content in fruits, may be indirectly influenced by CKs, as shown in kiwifruit [56] and strawberry [55]. Dipping application of kiwifruits in 10 mg/L CPPU significantly influenced the soluble carbohydrate component of the fruit osmotic pressure [56]. In apple, evidence of the expression of different genes related to CK activities was shown during axillary bud development [57] and flowering [58][59]. The expression of these CK-related genes was postulated to be essential for the postharvest storage of horticultural fruits, including strawberry [55]
. Gene expression-related responses to cytokinin application in different horticultural fruit crops
| Attribute and Fruit | Focus of Study | Response(s) | Reference | |
|---|---|---|---|---|
| Seed and Flowering | ||||
| San Pedro fig ‘Asteran’ Ficus carica L. |
Effect of CPPU on the parthenocarpy induction | CPPU upregulated phytohormone genes such as GA20ox, GA3ox, GID1, GID2, AUX/IAA, and GH3, while downregulating NCED, PP2C and ABF | [63] | [63 |
| Strawberry Fragaria vesca L. |
Effect of IAA, BA, ACC, and GA3 on FvPHL gene regulation during seedling development | BA, ACC and GA3 slightly regulated the gene expression of FvPHL3/5/6 by BA, FvPHL5 by ACC and GA3, and FvPHL3 by IAA, while ABA influenced the expression of all six FvPHL genes | [64] | [64 |
| Apple “Fuji’ Malus domestica Borkh. |
Effect of BA (5 mM), decapitation, and lovastatin on the expression of MdIPT and MdCKX genes in apple during axillary bud outgrowth | BA and decapitation treatment induced the upregulation of MdIPT, MdCKX, and MdPIN1 genes, while lovastatin (a compound that effectively suppresses axillary bud outgrowth) inhibited gene expression. Both BA and lovastatin upregulated MdCKX8 and MdCKX10 genes | [57] | [57 |
| Apple Malus domestica Borkh. |
Effect of 300 mg/L BA on floral genes (MdFT, AFL1, and MdTFL1) during flower development after 180 DAF | BA upregulated the transcription of MdFT at 110 DAF, AFL1 at 50 and 110 DAF, with a significant decline in MdTFL1 expression at 30 and 180 DAF | [58] | [58 |
| Fruit quality | ||||
| Apple “Pink Lady’ Malus x domestica Borkh. | Effect of GA4+7 and BA on the cellular mechanism of calyx-end cracking during fruit development | Early application of GA4+7 and BA (commercial product Superlon™ with 1.9% (v/v) of both plant growth hormone) increased epidermal cell density, which strengthened cell-wall components and upregulated the expression of genes responsible for fruit-cracking resistance | [65] | [65 |
| Biochemical and physiological parameters | ||||
| Kiwi ‘Hayward’ Actinidia chinensis var. deliciosa | Effect of CPPU on transcript abundance of carbohydrate metabolism genes at a standard and a high carbohydrate supply | CPPU-treated fruits reduced starch synthesis while increasing starch degradation during early fruit development (standard carbohydrate supply). However, CPPU-treated fruits increased vacuolar invertase transcripts, which in turn increased the sucrose cleavage associated with increased fructokinase (FK4) gene expression in early fruit development (high starch supply) | [56] | [56 |
| Kiwi ‘Xuxiang’ Actinidia deliciosa. | Effect of CPPU on volatile emissions and differential gene expression related to these compounds after days of storage | CPPU inhibited the biosynthesis of volatile compounds including aldehydes, esters, and terpenes. CPPU influenced gene expression related to hormone signal transduction in aldehydes, alcohols, and terpene biosynthetic pathways | [66] | [66 |
| Strawberry ‘Akihime’ Fragaria × ananassa Duch. |
Effect of CPPU application on the proteomic analysis during pre and postharvest | In total, 88 and 56 proteins were expressed during harvest and after storage, respectively. CPPU regulated glycolysis, photosynthesis, and acid metabolism before storage. Particularly, the upregulated expression of the LDOX gene contributed to the induction of anthocyanin content in strawberry in response to CPPU. However, CPPU suppressed volatile biosynthesis | [55] | [55 |
| Litchi ‘Feizixiao’ Litchi chinensis Sonn. |
Effect of 25 mg/L ABA and 4 mg/L CPPU on physiological changes and transcriptome profiling | ABA upregulated the expressions of genes (LcGST4) involved in flavonoid and anthocyanin biosynthesis, while CPPU induced genes related to carbon metabolism, amino acids, photosynthesis, and downregulated genes related to anthocyanin biosynthesis | [67] | [67 |
| Litchi ‘Feizixiao’ Litchi chinensis Sonn. |
Effect of ABA and CPPU on the expression of anthocyanin-related LcGST4 genes | ABA enhanced anthocyanin accumulation through the induced expression of LcGST4 during ripening stages. CPPU reduced anthocyanin production and LcGST4 expression remained at low levels | [68] | [68 |
| Pear ‘Cuiguan’ Pyrus pyrifolia Nakai | Effect of CPPU in verifying the function of B-PpRRs during fruit coloration and anthocyanin production in pear that never produce anthocyanin | CPPU stimulated anthocyanin production in the skin of fruitlets after 16 days of treatment. CPPU also induced B-PpRR anthocyanin biosynthetic genes, which are presumed to mediate anthocyanin production | [54] | [54 |
| Grape ‘Neo Muscat’ Vitis vinifera L. | Mutagenesis: expression of Vitis vinifera phytoene desaturase (VvPDS) gene | Carotenoid biosynthesis | [69] | [69 |
| Sweet orange ‘Valencia’ Citrus sinensis |
Mutagenesis: expression of Citrus sinensis Phytoene desaturase (CsPDS) gene | Carotenoid biosynthesis | [70] | [70 |
| Abiotic and biotic effects | ||||
| Wanjincheng orange Citrus sinensis Osbeck |
Mutagenesis: expression of Citrus sinensis Lateral organ boundaries 1 (CsLOB1) promoter | Citrus canker resistance | [71] | [71 |
| Duncan grapefruit Citrus paradisi | Mutagenesis: expression of CsLOB1 | Citrus canker resistance | [72] | [72 |
| Strawberry ‘Akihime’ Fragaria × ananassa Duch. |
Effect of CPPU application on the proteomic analysis during pre- and postharvest | In total, 88 and 56 proteins were expressed during harvest and after storage, respectively. CPPU application resulted in higher capacity of resistance in strawberry to stress stimuli after storage | [55] | [55 |
| Melon ‘Yangjiaomi’ Cucumis melo L. | Effect of tZ application on TCS genes | Type-A RRs, CmRRA1 - CmRRA7, were upregulated after 2 h of tZ application | [73] | [73 |
References
- Kaminek, M. Tracking the story of cytokinin research. J. Plant Growth Regul. 2015, 34, 723–739. [Google Scholar] [CrossRef]
- Merchant, S.S.; Gruissem, W.; Ort, D. Annual review of plant biology 2017. Curr. Sci. 2018, 115, 431–449. [Google Scholar] [CrossRef]
- Srivastava, L.M. Introduction to Some Special Aspects of Plant Growth and Development; Elsevier BV: Amsterdam, The Netherlands, 2002; p. 1. [Google Scholar]
- Stirk, W.A.; Van Staden, J. Flow of cytokinins through the environment. Plant Growth Regul. 2010, 62, 101–116. [Google Scholar] [CrossRef]
- Jameson, P.E.; Song, J. Cytokinin: A key driver of seed yield. J. Exp. Bot. 2015, 67, 593–606. [Google Scholar] [CrossRef]
- Kinet, J.; Lejeune, P.; Bernier, G. Shoot-root interactions during floral transition: A possible role for cytokinins. Environ. Exp. Bot. 1993, 33, 459–469. [Google Scholar] [CrossRef]
- Kumar, R.; Khurana, A.; Sharma, A.K. Role of plant hormones and their interplay in development and ripening of fleshy fruits. J. Exp. Bot. 2013, 65, 4561–4575. [Google Scholar] [CrossRef]
- Kunikowska, A.; Byczkowska, A.; Doniak, M.; Kaźmierczak, A. Cytokinins résumé: Their signaling and role in programmed cell death in plants. Plant Cell Rep. 2013, 32, 771–780. [Google Scholar] [CrossRef]
- Schmülling, T. New insights into the functions of cytokinins in plant development. J. Plant Growth Regul. 2002, 21, 40–49. [Google Scholar] [CrossRef]
- Šmehilová, M.; Spíchal, L. The biotechnological potential of cytokinin status manipulation. In The Plant Plasma Membrane; Springer Science and Business Media LLC: Berlin, Germany, 2013; Volume 22, pp. 103–130. [Google Scholar]
- Rademacher, W. Plant growth regulators: Backgrounds and uses in plant production. J. Plant Growth Regul. 2015, 34, 845–872. [Google Scholar] [CrossRef]
- Spíchal, L. Cytokinins—Recent news and views of evolutionally old molecules. Funct. Plant Biol. 2012, 39, 267. [Google Scholar] [CrossRef]
- Khush, G. Green revolution: The way forward. Nat. Rev. Genet. 2001, 2, 815–822. [Google Scholar] [CrossRef] [PubMed]
- Koprna, R.; De Diego, N.; Dundálková, L.; Spíchal, L. Use of cytokinins as agrochemicals. Bioorganic Med. Chem. 2016, 24, 484–492. [Google Scholar] [CrossRef] [PubMed]
- Bapat, V.A.; Trivedi, P.K.; Ghosh, A.; Sane, V.A.; Ganapathi, T.R.; Nath, P. Ripening of fleshy fruit: Molecular insight and the role of ethylene. Biotechnol. Adv. 2010, 28, 94–107. [Google Scholar] [CrossRef] [PubMed]
- Cherian, S.; Figueroa, C.R.; Nair, H. ‘Movers and shakers’ in the regulation of fruit ripening: A cross-dissection of climacteric versus non-climacteric fruit. J. Exp. Bot. 2014, 65, 4705–4722. [Google Scholar] [CrossRef]
- Debnath, S.C. Micropropagation of small fruits. In Somatic Embryogenesis in Woody Plants; Springer Science and Business Media LLC: Berlin, Germany, 2003; pp. 465–506. [Google Scholar]
- Nadeem, S.M.; Ahmad, M.; Zahir, Z.A.; Javaid, A.; Ashraf, M. The role of mycorrhizae and plant growth promoting rhizobacteria (PGPR) in improving crop productivity under stressful environments. Biotechnol. Adv. 2014, 32, 429–448. [Google Scholar] [CrossRef]
- Pauls, K. Plant biotechnology for crop improvement. Biotechnol. Adv. 1995, 13, 673–693. [Google Scholar] [CrossRef]
- Rai, M.K.; Asthana, P.; Singh, S.K.; Jaiswal, V.; Jaiswal, U. The encapsulation technology in fruit plants—A review. Biotechnol. Adv. 2009, 27, 671–679. [Google Scholar] [CrossRef]
- Sharma, H.; Crouch, J.; Sharma, K.; Seetharama, N.; Hash, C. Applications of biotechnology for crop improvement: Prospects and constraints. Plant Sci. 2002, 163, 381–395. [Google Scholar] [CrossRef]
- Zalabák, D.; Pospíšilová, H.; Šmehilová, M.; Mrízová, K.; Frébort, I.; Galuszka, P. Genetic engineering of cytokinin metabolism: Prospective way to improve agricultural traits of crop plants. Biotechnol. Adv. 2013, 31, 97–117. [Google Scholar] [CrossRef]
- Ma, Q.-H. Genetic engineering of cytokinins and their application to agriculture. Crit. Rev. Biotechnol. 2008, 28, 213–232. [Google Scholar] [CrossRef]
- Rai, M.K.; Shekhawat, N.S. Recent advances in genetic engineering for improvement of fruit crops. Plant Cell Tissue Organ Cult. 2013, 116, 1–15. [Google Scholar] [CrossRef]
- Sonah, H.; Deshmukh, R.; Singh, V.P.; Gupta, D.K.; Singh, N.K.; Sharma, T.R. Genomic resources in horticultural crops: Status, utility and challenges. Biotechnol. Adv. 2011, 29, 199–209. [Google Scholar] [CrossRef] [PubMed]
- Karkute, S.G.; Singh, A.K.; Gupta, O.P.; Singh, P.M.; Singh, B. CRISPR/Cas9 mediated genome engineering for improvement of horticultural crops. Front. Plant Sci. 2017, 8, 8. [Google Scholar] [CrossRef] [PubMed]
- Gururani, M.A.; Ganesan, M.; Song, P.-S. Photo-biotechnology as a tool to improve agronomic traits in crops. Biotechnol. Adv. 2015, 33, 53–63. [Google Scholar] [CrossRef]
- Manjunatha, G.; Lokesh, V.; Neelwarne, B. Nitric oxide in fruit ripening: Trends and opportunities. Biotechnol. Adv. 2010, 28, 489–499. [Google Scholar] [CrossRef]
- Watkins, C.B. The use of 1-methylcyclopropene (1-MCP) on fruits and vegetables. Biotechnol. Adv. 2006, 24, 389–409. [Google Scholar] [CrossRef]
- Sharma, H.S.S.; Fleming, C.; Selby, C.; Rao, J.R.; Martin, T. Plant biostimulants: A review on the processing of macroalgae and use of extracts for crop management to reduce abiotic and biotic stresses. Environ. Biol. Fishes 2013, 26, 465–490. [Google Scholar] [CrossRef]
- Albacete, A.; Martínez-Andújar, C.; Pérez-Alfocea, F. Hormonal and metabolic regulation of source-sink relations under salinity and drought: From plant survival to crop yield stability. Biotechnol. Adv. 2014, 32, 12–30. [Google Scholar] [CrossRef]
- Argueso, C.T.; Ferreira, F.J.; Kieber, J.J. Environmental perception avenues: The interaction of cytokinin and environmental response pathways. Plant Cell Environ. 2009, 32, 1147–1160. [Google Scholar] [CrossRef]
- Shigenaga, A.M.; Argueso, C.T. No hormone to rule them all: Interactions of plant hormones during the responses of plants to pathogens. Semin. Cell Dev. Biol. 2016, 56, 174–189. [Google Scholar] [CrossRef]
- Wolters, H.; Jürgens, G. Survival of the flexible: Hormonal growth control and adaptation in plant development. Nat. Rev. Genet. 2009, 10, 305–317. [Google Scholar] [CrossRef] [PubMed]
- Pacifici, E.; Polverari, L.; Sabatini, S. Plant hormone cross-talk: The pivot of root growth. J. Exp. Bot. 2015, 66, 1113–1121. [Google Scholar] [CrossRef] [PubMed]
- Vanková, R.; Petrášek, J.; Zažímalová, E.; Kamínek, M.; Motyka, V.; Ludwig-Müller, J. Auxins and cytokinins in plant development and interactions with other phytohormones 2014. J. Plant Growth Regul. 2014, 33, 709–714. [Google Scholar] [CrossRef]
- Bubán, T. The use of benzyladenine in orchard fruit growing: A mini review. Plant Growth Regul. 2000, 32, 381–390. [Google Scholar] [CrossRef]
- Kakimoto, T. Perception and signal transduction of cytokinins. Annu. Rev. Plant Biol. 2003, 54, 605–627. [Google Scholar] [CrossRef]
- Hwang, I.; Sheen, J.; Muller, B. Cytokinin signaling networks. Annu. Rev. Plant Biol. 2012, 63, 353–380. [Google Scholar] [CrossRef]
- Steklov, M.Y.; Lomin, S.N.; Osolodkin, D.I.; Romanov, G.A. Structural basis for cytokinin receptor signaling: An evolutionary approach. Plant Cell Rep. 2013, 32, 781–793. [Google Scholar] [CrossRef]
- Bergougnoux, V. The history of tomato: From domestication to biopharming. Biotechnol. Adv. 2014, 32, 170–189. [Google Scholar] [CrossRef]
- Gillaspy, G.E.; Ben-David, H.; Gruissem, W. Fruits: A developmental perspective. Plant Cell 1993, 5, 1439–1451. [Google Scholar] [CrossRef]
- Matsuo, S.; Kikuchi, K.; Fukuda, M.; Honda, I.; Imanishi, S. Roles and regulation of cytokinins in tomato fruit development. J. Exp. Bot. 2012, 63, 5569–5579. [Google Scholar] [CrossRef]
- Schaller, G.E.; Bishopp, A.; Kieber, J.J. The yin-yang of hormones: Cytokinin and auxin interactions in plant development. Plant Cell 2015, 27, 44–63. [Google Scholar] [CrossRef] [PubMed]
- Hosek, P.; Hoyerová, K.; Kiran, N.S.; Dobrev, P.I.; Zahajská, L.; Filepová, R.; Motyka, V.; Müller, K.; Kamínek, M. Distinct metabolism of N-glucosides of isopentenyladenine and trans-zeatin determines cytokinin metabolic spectrum in Arabidopsis. New Phytol. 2019, 225, 2423–2438. [Google Scholar] [CrossRef] [PubMed]
- Kurakawa, T.; Ueda, N.; Maekawa, M.; Kobayashi, K.; Kojima, M.; Nagato, Y.; Sakakibara, H.; Kyozuka, J. Direct control of shoot meristem activity by a cytokinin-activating enzyme. Nature 2007, 445, 652–655. [Google Scholar] [CrossRef] [PubMed]
- Kieber, J.J.; Schaller, G.E. Cytokinin signaling in plant development. Development 2018, 145, dev149344. [Google Scholar] [CrossRef]
- Kieber, J.J.; Schaller, G.E. Cytokinins. Arab. Book 2014, 12, e0168. [Google Scholar] [CrossRef]
- Šmehilová, M.; Dobrůšková, J.; Novak, O.; Takáč, T.; Galuszka, P. Cytokinin-specific glycosyltransferases possess different roles in cytokinin homeostasis maintenance. Front. Plant Sci. 2016, 7. [Google Scholar] [CrossRef]
- Argueso, C.T.; Ferreira, F.J.; Epple, P.; To, J.P.; Hutchison, C.E.; Schaller, G.E.; Dangl, J.L.; Kieber, J.J. Two-component elements mediate interactions between cytokinin and salicylic acid in plant immunity. PLoS Genet. 2012, 8, e1002448. [Google Scholar] [CrossRef]
- Shi, X.; Gupta, S.; Lindquist, I.E.; Cameron, C.T.; Mudge, J.; Rashotte, A.M. Transcriptome analysis of cytokinin response in tomato leaves. PLoS ONE 2013, 8, e55090. [Google Scholar] [CrossRef]
- Shi, X.; Gupta, S.; Rashotte, A.M. Characterization of two tomato AP2/ERF genes, SlCRF1 and SlCRF2 in hormone and stress responses. Plant Cell Rep. 2013, 33, 35–45. [Google Scholar] [CrossRef]
- Cortleven, A.; Leuendorf, J.E.; Frank, M.; Pezzetta, D.; Bolt, S.; Schmülling, T. Cytokinin action in response to abiotic and biotic stresses in plants. Plant Cell Environ. 2019, 42, 998–1018. [Google Scholar] [CrossRef]
- Ni, J.; Bai, S.; Gao, L.; Qian, M.; Zhong, L.; Teng, Y. Identification, classification, and transcription profiles of the B-type response regulator family in pear. PLoS ONE 2017, 12, e0171523. [Google Scholar] [CrossRef] [PubMed]
- Li, L.; Li, N.; Luo, Z.; Huang, X.; Li, X. Proteomic response and quality maintenance in postharvest fruit of strawberry (Fragaria × ananassa) to exogenous cytokinin. Sci. Rep. 2016, 6, 27094. [Google Scholar] [CrossRef] [PubMed]
- Nardozza, S.; Boldingh, H.L.; Wohlers, M.; Gleave, A.P.; Luo, Z.; Costa, G.; A Macrae, E.; Clearwater, M.J.; Richardson, A.C. Exogenous cytokinin application to Actinidia chinensis var. deliciosa ‘Hayward’ fruit promotes fruit expansion through water uptake. Hortic. Res. 2017, 4, 17043. [Google Scholar] [CrossRef] [PubMed]
- Tan, M.; Li, G.; Qi, S.; Liu, X.; Chen, X.; Ma, J.; Zhang, N.; Han, M. Identification and expression analysis of the IPT and CKX gene families during axillary bud outgrowth in apple (Malus domestica Borkh.). Gene 2018, 651, 106–117. [Google Scholar] [CrossRef]
- Li, Y.; Zhang, N.; Xing, L.; Zhang, S.; Zhao, C.; Han, M. Effect of exogenous 6-benzylaminopurine (6-BA) on branch type, floral induction and initiation, and related gene expression in ‘Fuji’ apple (Malus domestica Borkh.). Plant Growth Regul. 2015, 79, 65–70. [Google Scholar] [CrossRef]
- Li, Y.; Zhang, N.; Zhang, L.; Zuo, X.; Fan, S.; Zhang, X.; Shalmani, A.; Han, M. Identification and expression analysis of cytokinin response-regulator genes during floral induction in apple (Malus domestica Borkh.). Plant Growth Regul. 2017, 83, 455–464. [Google Scholar] [CrossRef]
- Tian, S.; Jiang, L.; Zhang, J.; Zong, M.; Ren, Y.; Guo, S.; Gong, G.; Liu, F.; Xu, Y.; Gao, Q. Efficient CRISPR/Cas9-based gene knockout in watermelon. Plant Cell Rep. 2016, 36, 399–406. [Google Scholar] [CrossRef]
- Hajari, E. Molecular and related approaches to litchi improvement—Historical perspective and future trends. J. Hortic. Sci. Biotechnol. 2019, 94, 693–702. [Google Scholar] [CrossRef]
- Wang, T.; Zhang, H.; Zhu, H.-L. CRISPR technology is revolutionizing the improvement of tomato and other fruit crops. Hortic. Res. 2019, 6, 77. [Google Scholar] [CrossRef]
- Chai, P.; Dong, S.; Chai, L.; Chen, S.; Flaishman, M.; Ma, H. Cytokinin-induced parthenocarpy of San Pedro type fig (Ficus carica L.) main crop: Explained by phytohormone assay and transcriptomic network comparison. Plant Mol. Biol. 2019, 99, 329–346. [Google Scholar] [CrossRef]
- Zheng, J.; Cao, M.; Zhang, Z.; Zheng, Z.-L. Expression analysis suggests potential roles for PH-LIKE(PHL) genes in diploid strawberry Fragaria vesca L. seedling hormone response and fruit development. J. Hortic. Sci. Biotechnol. 2018, 94, 151–159. [Google Scholar] [CrossRef]
- Joshi, M.; Baghel, R.S.; Fogelman, E.; Stern, R.A.; Ginzberg, I. Identification of candidate genes mediating apple fruit-cracking resistance following the application of gibberellic acids 4 + 7 and the cytokinin 6-benzyladenine. Plant Physiol. Biochem. 2018, 127, 436–445. [Google Scholar] [CrossRef] [PubMed]
- Luo, J.; Guo, L.; Huang, Y.; Wang, C.; Qiao, C.; Pang, R.; Li, J.; Pang, T.; Wang, R.; Xie, H.; et al. Transcriptome analysis reveals the effect of pre-harvest CPPU treatment on the volatile compounds emitted by kiwifruit stored at room temperature. Food Res. Int. 2017, 102, 666–673. [Google Scholar] [CrossRef] [PubMed]
- Hu, B.; Li, J.; Wang, D.; Qin, Y.; Zhao, J. Transcriptome profiling of Litchi chinensis pericarp in response to exogenous cytokinins and abscisic acid. Plant Growth Regul. 2017, 84, 437–450. [Google Scholar] [CrossRef]
- Hu, B.; Zhao, J.; Lai, B.; Qin, Y.; Wang, H.; Hu, G. LcGST4 is an anthocyanin-related glutathione S-transferase gene in Litchi chinensis Sonn. Plant Cell Rep. 2016, 35, 831–843. [Google Scholar] [CrossRef] [PubMed]
- Nakajima, I.; Ban, Y.; Azuma, A.; Onoue, N.; Moriguchi, T.; Yamamoto, T.; Toki, S.; Endo, M. CRISPR/Cas9-mediated targeted mutagenesis in grape. PLoS ONE 2017, 12, e0177966. [Google Scholar] [CrossRef] [PubMed]
- Jia, H.; Wang, N. Targeted genome editing of sweet orange using Cas9/sgRNA. PLoS ONE 2014, 9, e93806. [Google Scholar] [CrossRef]
- Peng, A.; Chen, S.; Lei, T.; Xu, L.; He, Y.; Wu, L.; Yao, L.; Zou, X. Engineering canker-resistant plants through CRISPR/Cas9-targeted editing of the susceptibility gene CsLOB1 promoter in citrus. Plant Biotechnol. J. 2017, 15, 1509–1519. [Google Scholar] [CrossRef]
- Jia, H.; Zhang, Y.; Orbović, V.; Xu, J.; White, F.F.; Jones, J.B.; Wang, N. Genome editing of the disease susceptibility gene CsLOB1 in citrus confers resistance to citrus canker. Plant Biotechnol. J. 2017, 15, 817–823. [Google Scholar] [CrossRef]
- Liu, P.; Wang, S.; Wang, X.; Yang, X.; Li, Q.; Wang, C.; Chen, C.; Shi, Q.; Ren, Z.; Wang, L. Genome-wide characterization of two-component system (TCS) genes in melon (Cucumis melo L.). Plant Physiol. Biochem. 2020, 151, 197–213. [Google Scholar] [CrossRef]
Recently, genome editing in fruit crops by CRISPR/Cas9 has emerged as an alternative approach to mitigate time-consuming conventional breeding programmes [26][60][61]. Since the first studies in tomato and citrus-producing stable transgenic lines, the CRISPR/Cas9 technology has been applied to an increasing list of fruit crops including kiwifruit, banana, strawberry, papaya, and ground berry [62]. Genome-wide expression analysis data are largely lacking for many aspects linked to the developmental biology of fruit crops. Available information is mainly for the fruit biology of horticultural crops and genes linked to defense responses but not necessarily linked to CK responsiveness [51]. Despite increasing efforts, the molecular mechanisms underlying the role of CKs in pre- and postharvest quality performance of horticultural fruits are yet to be fully elucidated, and such information may be critical for the utilisation of modern technologies for fruit crop improvement
References
- Kaminek, M. Tracking the story of cytokinin research. J. Plant Growth Regul. 2015, 34, 723–739, doi:10.1007/s00344-015-9543-4.
- Merchant, S.S.; Gruissem, W.; Ort, D. Annual review of plant biology 2017. Curr. Sci. 2018, 115, 431–449,, doi:10.18520/cs/v115/i6/1204-1207.
- Srivastava, L.M. Introduction to Some Special Aspects of Plant Growth and Development; Elsevier BV: Amsterdam, Netherlands, 2002; p. 1.
- Stirk, W.A.; Van Staden, J. Flow of cytokinins through the environment. Plant Growth Regul. 2010, 62, 101–116, doi:10.1007/s10725-010-9481-x.
- Jameson, P.E.; Song, J. Cytokinin: A key driver of seed yield. J. Exp. Bot. 2015, 67, 593–606, doi:10.1093/jxb/erv461.
- Kinet, J.; Lejeune, P.; Bernier, G. Shoot-root interactions during floral transition: A possible role for cytokinins. Environ. Exp. Bot. 1993, 33, 459–469, doi:10.1016/0098-8472(93)90019-c.
- Kumar, R.; Khurana, A.; Sharma, A.K. Role of plant hormones and their interplay in development and ripening of fleshy fruits. J. Exp. Bot. 2013, 65, 4561–4575, doi:10.1093/jxb/eru277.
- Kunikowska, A.; Byczkowska, A.; Doniak, M.; Kaźmierczak, A. Cytokinins résumé: Their signaling and role in programmed cell death in plants. Plant Cell Rep. 2013, 32, 771–780, doi:10.1007/s00299-013-1436-z.
- Schmülling, T. New insights into the functions of cytokinins in plant development. J. Plant Growth Regul. 2002, 21, 40–49, doi:10.1007/s003440010046.
- Šmehilová, M.; Spíchal, L. The biotechnological potential of cytokinin status manipulation. In The Plant Plasma Membrane; Springer Science and Business Media LLC: Berlin, Germany, 2013; Volume 22, pp. 103–130.
- Rademacher, W. Plant growth regulators: Backgrounds and uses in plant production. J. Plant Growth Regul. 2015, 34, 845–872, doi:10.1007/s00344-015-9541-6.
- Spíchal, L. Cytokinins—Recent news and views of evolutionally old molecules. Funct. Plant Biol. 2012, 39, 267, doi:10.1071/fp11276.
- Khush, G. Green revolution: The way forward. Nat. Rev. Genet. 2001, 2, 815–822, doi:10.1038/35093585.
- Koprna, R.; De Diego, N.; Dundálková, L.; Spíchal, L. Use of cytokinins as agrochemicals. Bioorganic Med. Chem. 2016, 24, 484–492, doi:10.1016/j.bmc.2015.12.022.
- Bapat, V.A.; Trivedi, P.K.; Ghosh, A.; Sane, V.A.; Ganapathi, T.R.; Nath, P. Ripening of fleshy fruit: Molecular insight and the role of ethylene. Biotechnol. Adv. 2010, 28, 94–107, doi:10.1016/j.biotechadv.2009.10.002.
- Cherian, S.; Figueroa, C.R.; Nair, H. ‘Movers and shakers’ in the regulation of fruit ripening: A cross-dissection of climacteric versus non-climacteric fruit. J. Exp. Bot. 2014, 65, 4705–4722, doi:10.1093/jxb/eru280.
- Debnath, S.C. Micropropagation of small fruits. In Somatic Embryogenesis in Woody Plants; Springer Science and Business Media LLC: Berlin, Germany, 2003; pp. 465–506.
- Nadeem, S.M.; Ahmad, M.; Zahir, Z.A.; Javaid, A.; Ashraf, M. The role of mycorrhizae and plant growth promoting rhizobacteria (PGPR) in improving crop productivity under stressful environments. Biotechnol. Adv. 2014, 32, 429–448, doi:10.1016/j.biotechadv.2013.12.005.
- Pauls, K. Plant biotechnology for crop improvement. Biotechnol. Adv. 1995, 13, 673–693, doi:10.1016/0734-9750(95)02010-1.
- Rai, M.K.; Asthana, P.; Singh, S.K.; Jaiswal, V.; Jaiswal, U. The encapsulation technology in fruit plants—A review. Biotechnol. Adv. 2009, 27, 671–679, doi:10.1016/j.biotechadv.2009.04.025.
- Sharma, H.; Crouch, J.; Sharma, K.; Seetharama, N.; Hash, C. Applications of biotechnology for crop improvement: Prospects and constraints. Plant Sci. 2002, 163, 381–395, doi:10.1016/s0168-9452(02)00133-4.
- Zalabák, D.; Pospíšilová, H.; Šmehilová, M.; Mrízová, K.; Frébort, I.; Galuszka, P. Genetic engineering of cytokinin metabolism: Prospective way to improve agricultural traits of crop plants. Biotechnol. Adv. 2013, 31, 97–117, doi:10.1016/j.biotechadv.2011.12.003.
- Ma, Q.-H. Genetic engineering of cytokinins and their application to agriculture. Crit. Rev. Biotechnol. 2008, 28, 213–232, doi:10.1080/07388550802262205.
- Rai, M.K.; Shekhawat, N.S. Recent advances in genetic engineering for improvement of fruit crops. Plant Cell Tissue Organ Cult. 2013, 116, 1–15, doi:10.1007/s11240-013-0389-9.
- Sonah, H.; Deshmukh, R.; Singh, V.P.; Gupta, D.K.; Singh, N.K.; Sharma, T.R. Genomic resources in horticultural crops: Status, utility and challenges. Biotechnol. Adv. 2011, 29, 199–209, doi:10.1016/j.biotechadv.2010.11.002.
- Karkute, S.G.; Singh, A.K.; Gupta, O.P.; Singh, P.M.; Singh, B. CRISPR/Cas9 mediated genome engineering for improvement of horticultural crops. Front. Plant Sci. 2017, 8, 8,, doi:10.3389/fpls.2017.01635.
- Gururani, M.A.; Ganesan, M.; Song, P.-S. Photo-biotechnology as a tool to improve agronomic traits in crops. Biotechnol. Adv. 2015, 33, 53–63, doi:10.1016/j.biotechadv.2014.12.005.
- Manjunatha, G.; Lokesh, V.; Neelwarne, B. Nitric oxide in fruit ripening: Trends and opportunities. Biotechnol. Adv. 2010, 28, 489–499, doi:10.1016/j.biotechadv.2010.03.001.
- Watkins, C.B. The use of 1-methylcyclopropene (1-MCP) on fruits and vegetables. Biotechnol. Adv. 2006, 24, 389–409, doi:10.1016/j.biotechadv.2006.01.005.
- Sharma, H.S.S.; Fleming, C.; Selby, C.; Rao, J.R.; Martin, T. Plant biostimulants: A review on the processing of macroalgae and use of extracts for crop management to reduce abiotic and biotic stresses. Environ. Biol. Fishes 2013, 26, 465–490, doi:10.1007/s10811-013-0101-9.
- Albacete, A.; Martínez-Andújar, C.; Pérez-Alfocea, F. Hormonal and metabolic regulation of source-sink relations under salinity and drought: From plant survival to crop yield stability. Biotechnol. Adv. 2014, 32, 12–30, doi:10.1016/j.biotechadv.2013.10.005.
- Argueso, C.T.; Ferreira, F.J.; Kieber, J.J. Environmental perception avenues: The interaction of cytokinin and environmental response pathways. Plant Cell Environ. 2009, 32, 1147–1160, doi:10.1111/j.1365-3040.2009.01940.x.
- Shigenaga, A.M.; Argueso, C.T. No hormone to rule them all: Interactions of plant hormones during the responses of plants to pathogens. Semin. Cell Dev. Biol. 2016, 56, 174–189, doi:10.1016/j.semcdb.2016.06.005.
- Wolters, H.; Jürgens, G. Survival of the flexible: Hormonal growth control and adaptation in plant development. Nat. Rev. Genet. 2009, 10, 305–317, doi:10.1038/nrg2558.
- Pacifici, E.; Polverari, L.; Sabatini, S. Plant hormone cross-talk: The pivot of root growth. J. Exp. Bot. 2015, 66, 1113–1121, doi:10.1093/jxb/eru534.
- Vanková, R.; Petrášek, J.; Zažímalová, E.; Kamínek, M.; Motyka, V.; Ludwig-Müller, J. Auxins and cytokinins in plant development and interactions with other phytohormones 2014. J. Plant Growth Regul. 2014, 33, 709–714, doi:10.1007/s00344-014-9449-6.
- Bubán, T. The use of benzyladenine in orchard fruit growing: A mini review. Plant Growth Regul. 2000, 32, 381–390, doi:10.1023/a:1010785604339.
- Kakimoto, T. Perception and signal transduction of cytokinins. Annu. Rev. Plant Biol. 2003, 54, 605–627, doi:10.1146/annurev.arplant.54.031902.134802.
- Hwang, I.; Sheen, J.; Muller, B. Cytokinin signaling networks. Annu. Rev. Plant Biol. 2012, 63, 353–380, doi:10.1146/annurev-arplant-042811-105503.
- Steklov, M.Y.; Lomin, S.N.; Osolodkin, D.I.; Romanov, G.A. Structural basis for cytokinin receptor signaling: An evolutionary approach. Plant Cell Rep. 2013, 32, 781–793, doi:10.1007/s00299-013-1408-3.
- Bergougnoux, V. The history of tomato: From domestication to biopharming. Biotechnol. Adv. 2014, 32, 170–189, doi:10.1016/j.biotechadv.2013.11.003.
- Gillaspy, G.E.; Ben-David, H.; Gruissem, W. Fruits: A developmental perspective. Plant Cell 1993, 5, 1439–1451, doi:10.1105/tpc.5.10.1439.
- Matsuo, S.; Kikuchi, K.; Fukuda, M.; Honda, I.; Imanishi, S. Roles and regulation of cytokinins in tomato fruit development. J. Exp. Bot. 2012, 63, 5569–5579, doi:10.1093/jxb/ers207.
- Schaller, G.E.; Bishopp, A.; Kieber, J.J. The yin-yang of hormones: Cytokinin and auxin interactions in plant development. Plant Cell 2015, 27, 44–63, doi:10.1105/tpc.114.133595.
- Hosek, P.; Hoyerová, K.; Kiran, N.S.; Dobrev, P.I.; Zahajská, L.; Filepová, R.; Motyka, V.; Müller, K.; Kamínek, M. Distinct metabolism of N‐glucosides of isopentenyladenine and trans‐zeatin determines cytokinin metabolic spectrum in Arabidopsis. New Phytol. 2019, 225, 2423–2438, doi:10.1111/nph.16310.
- Kurakawa, T.; Ueda, N.; Maekawa, M.; Kobayashi, K.; Kojima, M.; Nagato, Y.; Sakakibara, H.; Kyozuka, J. Direct control of shoot meristem activity by a cytokinin-activating enzyme. Nature 2007, 445, 652–655, doi:10.1038/nature05504.
- Kieber, J.J.; Schaller, G.E. Cytokinin signaling in plant development. Development 2018, 145, dev149344, doi:10.1242/dev.149344.
- Kieber, J.J.; Schaller, G.E. Cytokinins. Arab. Book 2014, 12, e0168, doi:10.1199/tab.0168.
- Šmehilová, M.; Dobrůšková, J.; Novak, O.; Takáč, T.; Galuszka, P. Cytokinin-specific glycosyltransferases possess different roles in cytokinin homeostasis maintenance. Front. Plant Sci. 2016, 7, doi:10.3389/fpls.2016.01264.
- Argueso, C.T.; Ferreira, F.J.; Epple, P.; To, J.P.; Hutchison, C.E.; Schaller, G.E.; Dangl, J.L.; Kieber, J.J. Two-component elements mediate interactions between cytokinin and salicylic acid in plant immunity. PLoS Genet. 2012, 8, e1002448, doi:10.1371/journal.pgen.1002448.
- Shi, X.; Gupta, S.; Lindquist, I.E.; Cameron, C.T.; Mudge, J.; Rashotte, A.M. Transcriptome analysis of cytokinin response in tomato leaves. PLoS ONE 2013, 8, e55090, doi:10.1371/journal.pone.0055090.
- Shi, X.; Gupta, S.; Rashotte, A.M. Characterization of two tomato AP2/ERF genes, SlCRF1 and SlCRF2 in hormone and stress responses. Plant Cell Rep. 2013, 33, 35–45, doi:10.1007/s00299-013-1510-6.
- Cortleven, A.; Leuendorf, J.E.; Frank, M.; Pezzetta, D.; Bolt, S.; Schmülling, T. Cytokinin action in response to abiotic and biotic stresses in plants. Plant Cell Environ. 2019, 42, 998–1018, doi:10.1111/pce.13494.
- Ni, J.; Bai, S.; Gao, L.; Qian, M.; Zhong, L.; Teng, Y. Identification, classification, and transcription profiles of the B-type response regulator family in pear. PLoS ONE 2017, 12, e0171523, doi:10.1371/journal.pone.0171523.
- Li, L.; Li, N.; Luo, Z.; Huang, X.; Li, X. Proteomic response and quality maintenance in postharvest fruit of strawberry (Fragaria × ananassa) to exogenous cytokinin. Sci. Rep. 2016, 6, 27094, doi:10.1038/srep27094.
- Nardozza, S.; Boldingh, H.L.; Wohlers, M.; Gleave, A.P.; Luo, Z.; Costa, G.; A Macrae, E.; Clearwater, M.J.; Richardson, A.C. Exogenous cytokinin application to Actinidia chinensis var. deliciosa ‘Hayward’ fruit promotes fruit expansion through water uptake. Hortic. Res. 2017, 4, 17043, doi:10.1038/hortres.2017.43.
- Tan, M.; Li, G.; Qi, S.; Liu, X.; Chen, X.; Ma, J.; Zhang, N.; Han, M. Identification and expression analysis of the IPT and CKX gene families during axillary bud outgrowth in apple (Malus domestica Borkh.). Gene 2018, 651, 106–117, doi:10.1016/j.gene.2018.01.101.
- Li, Y.; Zhang, N.; Xing, L.; Zhang, S.; Zhao, C.; Han, M. Effect of exogenous 6-benzylaminopurine (6-BA) on branch type, floral induction and initiation, and related gene expression in ‘Fuji’ apple (Malus domestica Borkh.). Plant Growth Regul. 2015, 79, 65–70, doi:10.1007/s10725-015-0111-5.
- Li, Y.; Zhang, N.; Zhang, L.; Zuo, X.; Fan, S.; Zhang, X.; Shalmani, A.; Han, M. Identification and expression analysis of cytokinin response-regulator genes during floral induction in apple (Malus domestica Borkh.). Plant Growth Regul. 2017, 83, 455–464, doi:10.1007/s10725-017-0311-2.
- Tian, S.; Jiang, L.; Zhang, J.; Zong, M.; Ren, Y.; Guo, S.; Gong, G.; Liu, F.; Xu, Y.; Gao, Q. Efficient CRISPR/Cas9-based gene knockout in watermelon. Plant Cell Rep. 2016, 36, 399–406, doi:10.1007/s00299-016-2089-5.
- Hajari, E. Molecular and related approaches to litchi improvement—Historical perspective and future trends. J. Hortic. Sci. Biotechnol. 2019, 94, 693–702, doi:10.1080/14620316.2019.1624202.
- Wang, T.; Zhang, H.; Zhu, H.-L. CRISPR technology is revolutionizing the improvement of tomato and other fruit crops. Hortic. Res. 2019, 6, 77, doi:10.1038/s41438-019-0159-x.
- Chai, P.; Dong, S.; Chai, L.; Chen, S.; Flaishman, M.; Ma, H. Cytokinin-induced parthenocarpy of San Pedro type fig (Ficus carica L.) main crop: Explained by phytohormone assay and transcriptomic network comparison. Plant Mol. Biol. 2019, 99, 329–346, doi:10.1007/s11103-019-00820-2.
- Zheng, J.; Cao, M.; Zhang, Z.; Zheng, Z.-L. Expression analysis suggests potential roles for PH-LIKE(PHL) genes in diploid strawberry Fragaria vesca L. seedling hormone response and fruit development. J. Hortic. Sci. Biotechnol. 2018, 94, 151–159, doi:10.1080/14620316.2018.1499424.
- Joshi, M.; Baghel, R.S.; Fogelman, E.; Stern, R.A.; Ginzberg, I. Identification of candidate genes mediating apple fruit-cracking resistance following the application of gibberellic acids 4 + 7 and the cytokinin 6-benzyladenine. Plant Physiol. Biochem. 2018, 127, 436–445, doi:10.1016/j.plaphy.2018.04.015.
- Luo, J.; Guo, L.; Huang, Y.; Wang, C.; Qiao, C.; Pang, R.; Li, J.; Pang, T.; Wang, R.; Xie, H.; et al. Transcriptome analysis reveals the effect of pre-harvest CPPU treatment on the volatile compounds emitted by kiwifruit stored at room temperature. Food Res. Int. 2017, 102, 666–673, doi:10.1016/j.foodres.2017.09.051.
- Hu, B.; Li, J.; Wang, D.; Qin, Y.; Zhao, J. Transcriptome profiling of Litchi chinensis pericarp in response to exogenous cytokinins and abscisic acid. Plant Growth Regul. 2017, 84, 437–450, doi:10.1007/s10725-017-0351-7.
- Hu, B.; Zhao, J.; Lai, B.; Qin, Y.; Wang, H.; Hu, G. LcGST4 is an anthocyanin-related glutathione S-transferase gene in Litchi chinensis Sonn. Plant Cell Rep. 2016, 35, 831–843, doi:10.1007/s00299-015-1924-4.
- Nakajima, I.; Ban, Y.; Azuma, A.; Onoue, N.; Moriguchi, T.; Yamamoto, T.; Toki, S.; Endo, M. CRISPR/Cas9-mediated targeted mutagenesis in grape. PLoS ONE 2017, 12, e0177966, doi:10.1371/journal.pone.0177966.
- Jia, H.; Wang, N. Targeted genome editing of sweet orange using Cas9/sgRNA. PLoS ONE 2014, 9, e93806, doi:10.1371/journal.pone.0093806.
- Peng, A.; Chen, S.; Lei, T.; Xu, L.; He, Y.; Wu, L.; Yao, L.; Zou, X. Engineering canker-resistant plants through CRISPR/Cas9-targeted editing of the susceptibility gene CsLOB1 promoter in citrus. Plant Biotechnol. J. 2017, 15, 1509–1519, doi:10.1111/pbi.12733.
- Jia, H.; Zhang, Y.; Orbović, V.; Xu, J.; White, F.F.; Jones, J.B.; Wang, N. Genome editing of the disease susceptibility gene CsLOB1 in citrus confers resistance to citrus canker. Plant Biotechnol. J. 2017, 15, 817–823, doi:10.1111/pbi.12677.
- Liu, P.; Wang, S.; Wang, X.; Yang, X.; Li, Q.; Wang, C.; Chen, C.; Shi, Q.; Ren, Z.; Wang, L. Genome-wide characterization of two-component system (TCS) genes in melon (Cucumis melo L.). Plant Physiol. Biochem. 2020, 151, 197–213, doi:10.1016/j.plaphy.2020.03.017.
