Arginine is a semi-essential amino acid with a complex metabolism pathway and diverse functionality, which spans well beyond its role in the immune system. Cancers that are ‘arginine auxotrophic’ rely on extracellular arginine as a crucial substrate for proliferation and growth. Capitalizing on this vulnerability, there are numerous clinical trials evaluating the therapeutic benefits of depleting arginine in multiple types of cancer, including those occurring in the head and neck. However, head and neck cancers are different and are nonauxotrophic for arginine. Here, wresearchers explored the intricacies of arginine metabolism in tongue cancer in order to better understand the therapeutic potential of this biological vulnerability.
- oral tongue squamous cancer
- head and neck cancer
- arginase 1
- arginine metabolism
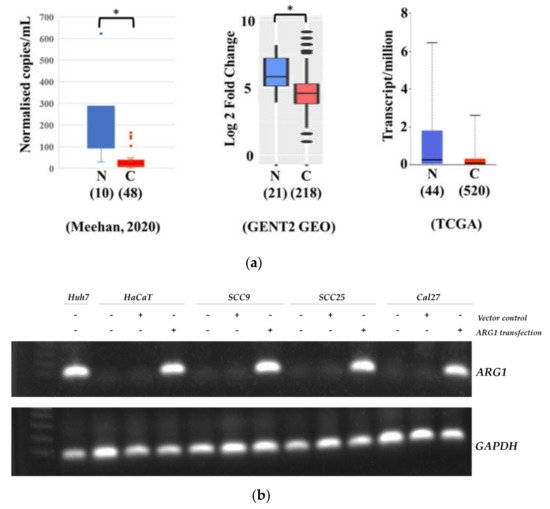
1. Gene and Protein Expression of ARG1 in Oral Squamous Cell Carcinoma Cell Lines
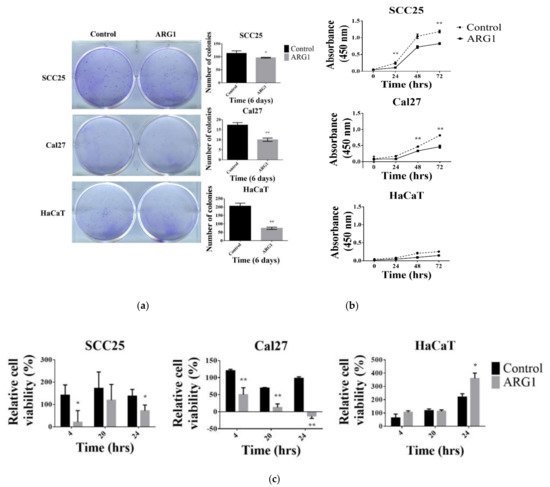
2. ARG1 Expression Inhibits Oral Squamous Cell Carcinoma Cell Growth
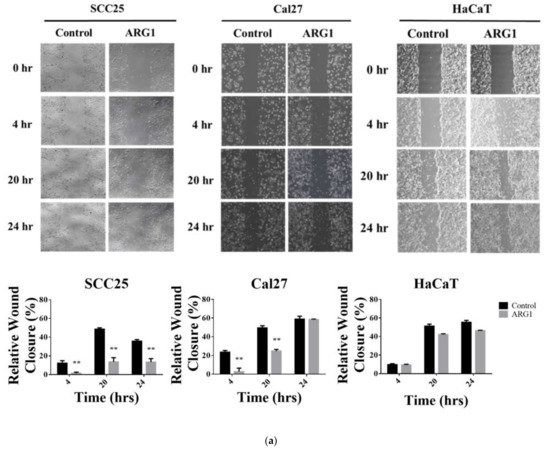
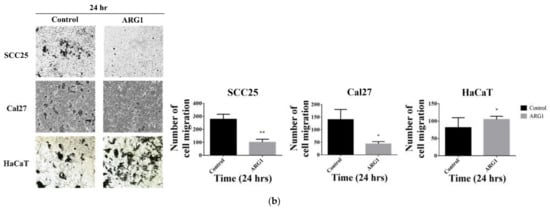
3. ARG1 Expression Inhibits Oral Squamous Cell Carcinoma Cell Migration and Invasion
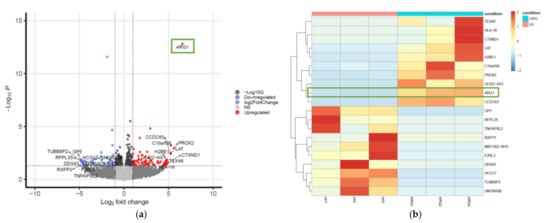
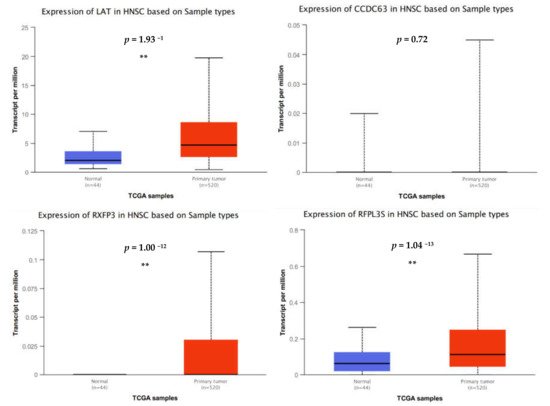
4. Transcriptome Profiling in SCC25 Delineates the Antioncogenic Effect of ARG1-Treated Oral Squamous Cell Carcinoma Cells
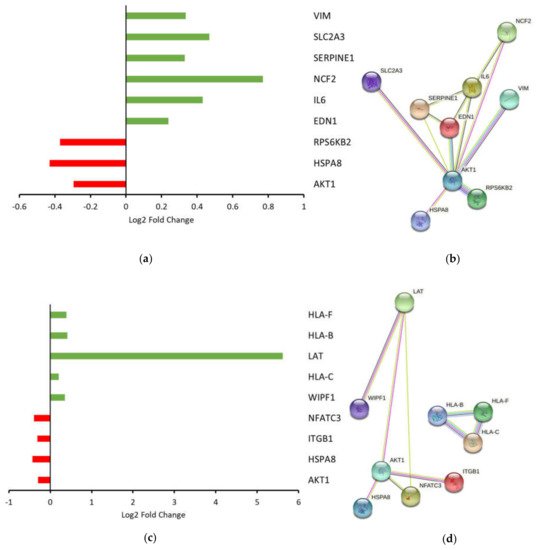
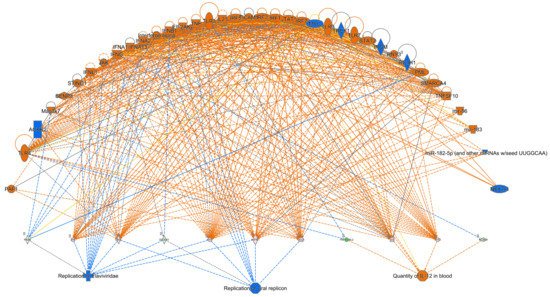
5. Ingenuity Pathway Analysis (IPA)
References
- Srivastava, S.; Ghosh, S.K. Modulation of L-Arginine-Arginase Metabolic Pathway Enzymes: Immunocytochemistry and mRNA Expression in Peripheral Blood and Tissue Levels in Head and Neck Squamous Cell Carcinomas in North East India. Asian Pac. J. Cancer Prev. 2015, 16, 7031–7038.
- Meehan, K.; Leslie, C.; Lucas, M.; Jacques, A.; Mirzai, B.; Lim, J.; Bulsara, M.; Khan, Y.; Wong, N.C.; Solomon, B.; et al. Characterization of the immune profile of oral tongue squamous cell carcinomas with advancing disease. Cancer Med. 2020, 9, 4791–4807.
- Chandrashekar, D.S.; Bashel, B.; Balasubramanya, S.A.H.; Creighton, C.J.; Ponce-Rodriguez, I.; Chakravarthi, B.; Varambally, S. UALCAN: A Portal for Facilitating Tumor Subgroup Gene Expression and Survival Analyses. Neoplasia 2017, 19, 649–658.
- Park, S.J.; Yoon, B.H.; Kim, S.K.; Kim, S.Y. GENT2: An updated gene expression database for normal and tumor tissues. BMC Med. Genom. 2019, 12, 101.
- Vishnubalaji, R.; Sasidharan Nair, V.; Ouararhni, K.; Elkord, E.; Alajez, N.M. Integrated Transcriptome and Pathway Analyses Revealed Multiple Activated Pathways in Breast Cancer. Front. Oncol. 2019, 9, 910.
