The modification and degradation of pectin in cell walls are necessary for the fruit softening process, which involves a series of pectin-modifying enzymes. Polygalacturonases (PGs) are a major group of pectin-hydrolyzing enzymes, which participate in fruit maturation, organ shedding, pollen development, and other processes by catalyzing the degradation of polygalacturonic acid. However, their function in plants has not yet been fully elucidated. In this paper, a full-length cDNA encoding SlPG49 was cloned from a tomato. Sequence alignment and phylogenetic analysis demonstrated that SlPG49 contains four typical conserved domains and belongs to clade E in PG classification. Quantitative real-time PCR analysis showed that SlPG49 was highly expressed in fruits during the softening stage, indicating that SlPG49 may be involved in fruit softening. Subcellular localization results revealed that SlPG49 was located in the cell membrane and the cell wall. In addition, an in vitro enzymatic activity assay confirmed that SlPG49 does have the ability to catalyze the hydrolysis of polygalacturonic acid. These results indicate that SlPG49 is a newly discovered PG gene involved in tomato fruit softening, and provide an experimental basis for elucidating the biological functions of plant PGs during fruit softening.
- polygalacturonases (PG)
- Solanum lycopersicum
- SlPG49
- gene expression
- enzymatic activity
1. Analysis on Research Results
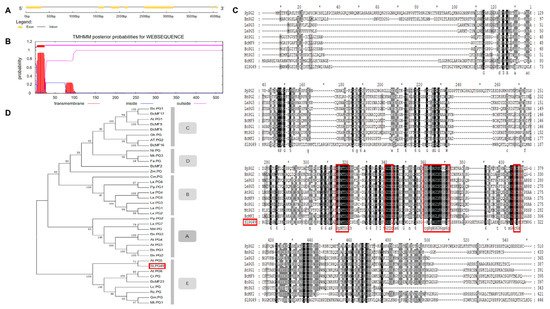
1.1. Sequence and Phylogenetic Analysis of SlPG49
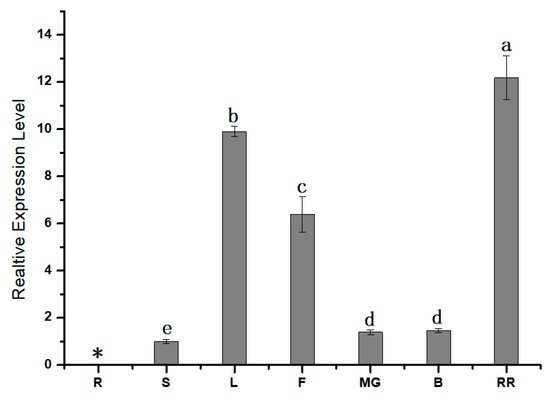
1.2. Expression Pattern Analysis of SlPG49
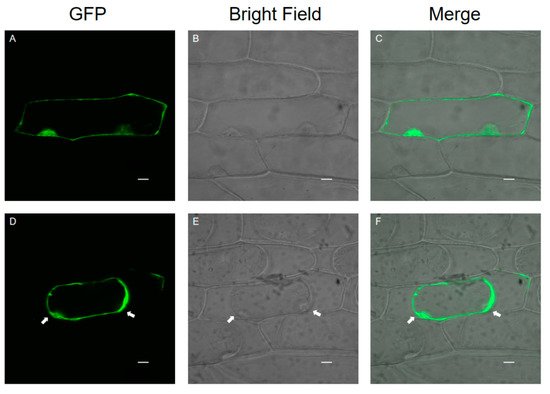
1.3. Subcellular Localization of SlPG49
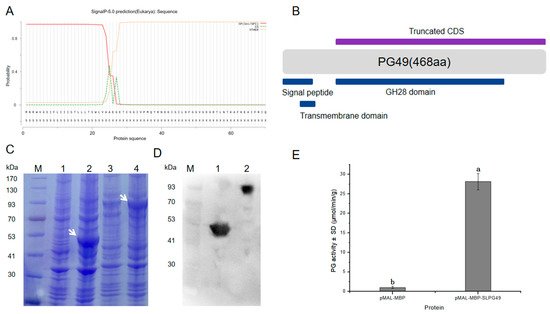
1.4. Enzymatic Activity of SlPG49 In Vitro
2. Current Insights
References
- Liang, Y.; Yu, Y.; Cui, J.; Lyu, M.; Xu, L.; Cao, J. A comparative analysis of the evolution, expression, and cis-regulatory element of polygalacturonase genes in grasses and dicots. Funct. Integr. Genom. 2016, 16, 641–656.
- Hocq, L.; Guinand, S.; Habrylo, O.; Voxeur, A.; Tabi, W.; Safran, J.; Fournet, F.; Domon, J.M.; Mollet, J.C.; Pilard, S.; et al. The exogenous application of AtPGLR, an endo-polygalacturonase, triggers pollen tube burst and repair. Plant J. 2020, 103, 617–633.
- Park, K.C.; Kwon, S.J.; Kim, P.H.; Bureau, T.; Kim, N.S. Gene structure dynamics and divergence of the polygalacturonase gene family of plants and fungus. Genome 2008, 51, 30–40.
- Bussink, H.J.D.; Buxton, F.P.; Visse, J. Expression and sequence comparison of the Aspergillus-niger and Aspergillus-tubigensis genes encoding polygalacturonase-II. Curr. Genet. 1991, 19, 467–474.
- Prasanna, V.; Prabha, T.N.; Tharanathan, R.N. Fruit Ripening Phenomena-An Overview. Crit. Rev. Food Sci. 2007, 47, 1–19.
- Ke, X.; Wang, H.; Li, Y.; Zhu, B.; Zang, Y.; He, Y.; Cao, J.; Zhu, Z.; Yu, Y. Genome-Wide identification and analysis of polygalacturonase genes in Solanum lycopersicum. Int. J. Mol. Sci. 2018, 19, 2290.
- Asl, L.K.; Dhondt, S.; Boudolf, V.; Beemster, G.T.S.; Beeckman, T.; Inzé, D.; Govaerts, W.; De Veylder, L. Model-based analysis of Arabidopsis leaf epidermal cells reveals distinct division and expansion patterns for pavement and guard cell. Plant Physiol. 2011, 156, 2172–2183.
- Caroli, M.D.; Lenucci, M.S.; Sansebastiano, G.P.D.; Dalessandro, G.; Lorenzo, G.D.; Piro, G. Protein trafficking to the cell wall occurs through mechanisms distinguishable from default sorting in tobacco. Plant J. 2011, 65, 295–308.
- Caroli, M.D.; Lenucci, M.S.; Sansebastiano, G.P.D.; Dalessandro, G.; Lorenzo, G.D.; Piro, G. Dynamic protein trafficking to the cell wall. Plant Signal. Behav. 2011, 6, 1012–1015.
- Lin, S.; Huang, L.; Yu, X.; Xiong, X.; Yue, X.; Liu, T.; Liang, Y.; Lv, M.; Cao, J. Characterization of BcMF23a and BcMF23b, two putative pectin methylesterase genes related to pollen development in Brassica campestris ssp. chinensis. Mol. Biol. Rep. 2017, 44, 139–148.
- Yang, Y.; Anderson, C.T.; Cao, J. Polygalacturonase45 cleaves pectin and links cell proliferation and morphogenesis to leaf curvature in Arabidopsis thaliana. Plant J. 2021, 106, 1493–1508.
- Rui, Y.; Xiao, C.; Yi, H.; Kandemir, B.; Wang, J.Z.; Puri, V.M.; Anderson, C.T. POLYGALACTURONASE INVOLVED IN EXPANSION3 functions in seedling development, rosette growth, and stomatal dynamics in Arabidopsis thaliana. Plant Cell 2017, 29, 2413–2432.
- Babu, Y.; Musielak, T.; Henschen, A.; Bayer, M. Suspensor length determines developmental progression of the embryo in Arabidopsis. Plant Physiol. 2013, 162, 1448–1458.
- Rhee, S.Y.; Osborne, E.; Poindexter, P.D.; Somerville, C.R. Microspore separation in the quartet 3 mutants of Arabidopsis is impaired by a defect in a developmentally regulated polygalacturonase required for pollen mother cell wall degradation. Plant Physiol. 2003, 133, 1170–1180.
- Ogawa, M.; Kay, P.; Wilson, S.; Swain, S.M. Arabidopsis dehiscence zone polygalacturonase1 (ADPG1), ADPG2, and QUARTET2 are polygalacturonases required for cell separation during reproductive development in Arabidopsis. Plant Cell 2009, 21, 216–233.
- Posé, S.; Kirby, A.R.; Paniagua, C.; Waldron, K.W.; Morris, V.J.; Quesada, M.A.; Mercado, J.A. The nanostructural characterization of strawberry pectins in pectate lyase or polygalacturonase silenced fruits elucidates their role in softening. Carbohydr. Polym. 2015, 132, 134–145.
- Atkinson, R.G.; Sutherland, P.W.; Johnston, S.L.; Gunaseelan, K.; Hallett, I.C.; Mitra, D.; Brummell, D.A.; Schröder, R.; Johnston, J.W.; Schaffer, R.J. Down-regulation of POLYGALACTURONASE1 alters firmness, tensile strength and water loss in apple (Malus × domestica) fruit. BMC Plant Biol. 2012, 12, 129.
- Asif, M.H.; Nath, P. Expression of multiple forms of polygalacturonase gene during ripening in banana fruit. Plant Physiol. Biochem. 2005, 43, 177–184.
