Multidrug and toxic compound extrusion (MATE) is one of the five major transporter superfamilies in living organisms. MATEs are characterized by 12 transmembrane α-helical domains, which are distinct from other solute transporters. In Glycine max, there are 117 MATE transporters gene family members. GmMATE1 and GmMATE2 are reported to mediate the accumulation of isoflavones in soybean seeds.
- soybean
- seed
- isoflavone
- multidrug and toxic compound extrusion (MATE) transporter
- genistein
- daidzein
- glycitein
1.GmMATE1 and GmMATE2 Are Isoflavone Transporters in Soybeans
GmMATE1W05 and GmMATE1C08 differ in two non-synonymous single-nucleotide polymorphisms (SNPs) while GmMATE2W05 and GmMATE2C08 differ in one non-synonymous SNP[1]. Transgenic yeasts expressing GmMATE1C08, GmMATE1W05, GmMATE2C08, GmMATE2W05 or the empty vector control were fed with isoflavone aglycones (a mixture of genistein, daidzein, and glycintein) (Figure 1). The isoflavones in the cells were then extracted and quantitated by UPLC. When fed with a mixture of genistein, daidzein, and glycitein, the aglycone form of these isoflavones, the transgenic yeasts expressing GmMATE1 and GmMATE2 significantly accumulated more of all three metabolites compared to the yeast transformed with the empty vector (EV) (Figure 1). The allelic forms from either the cultivated soybean C08 or the wild soybean W05 exhibited similar activities.

Figure 1. Yeast uptake assay using a mixture of genistein, daidzein, and glycitein. The level of (A) genistein, (B) daidzein, (C) glycitein, or (D) total aglycone (genistein, daidzein and glycitein) was determined. Metabolites were extracted from the yeast after feeding for 24 h and analyzed using UPLC against the respective standards. Values obtained from the yeast expressing GmMATE1 or GmMATE2 were normalized to those from the empty vector control (EV) in the same experiment. Values shown were the mean of six independent experiments ± SEM. Significant differences compared to EV were determined by Mann-Whitney test. *, p < 0.05; **, p < 0.01.
When the yeast constructs were fed with a mixture of isoflavone glycosides (genistin, daidzin, and glycitin), yeasts expressing GmMATE1C08 or GmMATE1W05 were able to accumulate more aglycones as well as more daidzin compared to the EV-transformed yeasts. Both aglycone and glycoside forms were detected in the metabolite extracts, probably due to the activities of endogenous hydrolytic enzymes which cleaved the sugar from the glycosides in the yeast cells (Figure 2). In the same experiment, although the yeasts expressing GmMATE2 also showed a similar trend in secondary metabolite accumulation to GmMATE1-expressing yeasts, the improved accumulation was not statistically significant, suggesting that GmMATE1 may have a higher activity than GmMATE2.
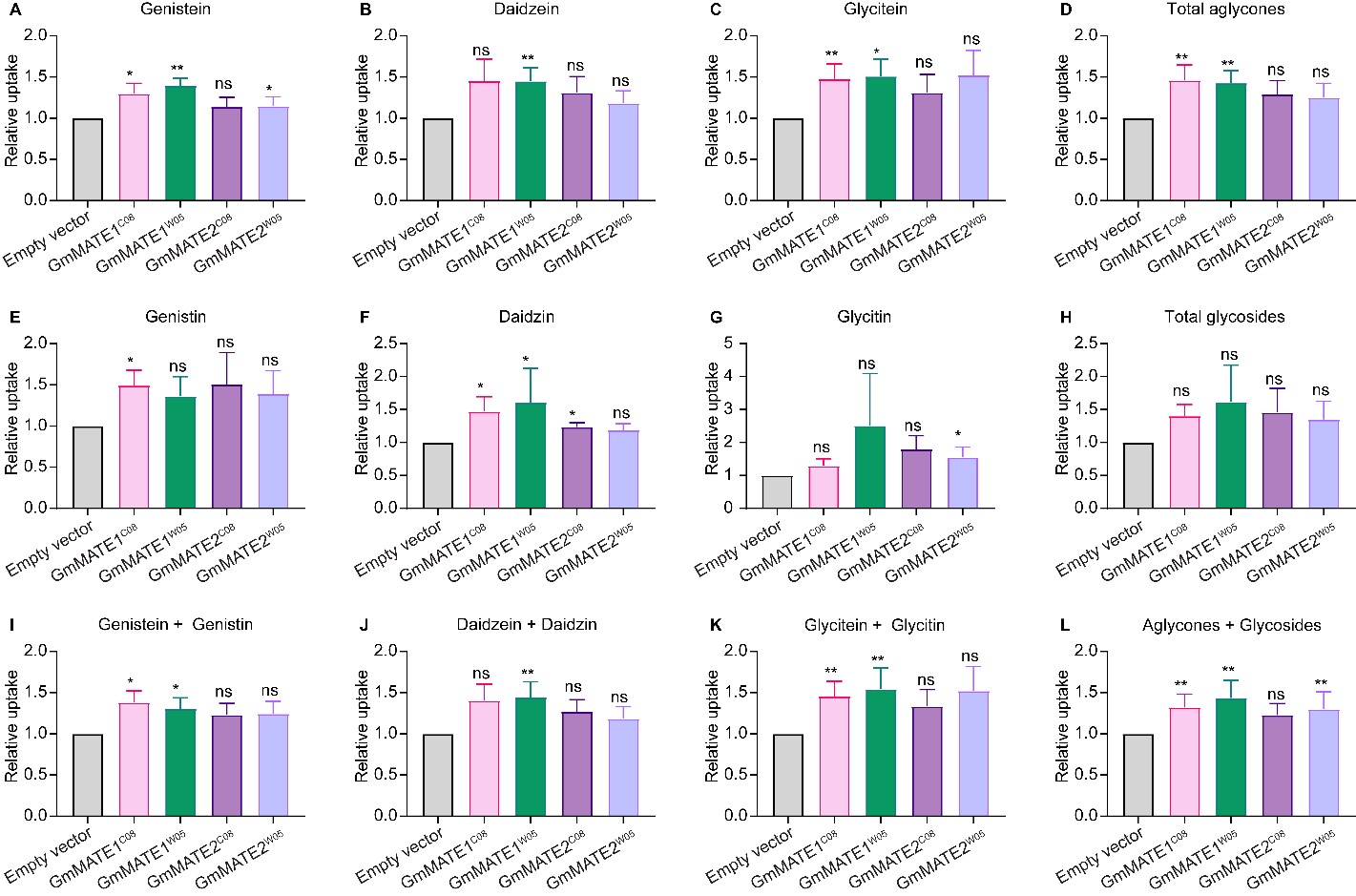
Figure 2. Yeast uptake assay with a mixture of genistin, daidzin, and glycitin. Metabolites were extracted from the yeast after feeding for 24 h and analyzed using UPLC against the respective standards. It appeared some of the glycosides were converted to aglycones in the yeast by endogenous enzymes. Values obtained from the yeast expressing GmMATEs were normalized to those from the empty vector control (EV) in the same experiment. The relative uptake levels of individual isoflavone species (A–C,E–G) and different isoflavone combinations (D,H,I–L) were determined. Total aglycones (D) refers to the combination of genistein, daidzein, and glyciteins. Total glycosides (H) refers to the combination of genistin, daidzin, and glycitin. The relative uptake level of total aglycones and glycosides is shown in (L). Values shown are the mean of six independent experiments ± SEM. Significant differences compared to EV were determined by Mann-Whitney test. *, p < 0.05; **, p < 0.01; ns, not significant.
2. Both GmMATE1 and GmMATE2 Were Localized in the Vacuolar Membrane
A subcellular localization study was conducted using transgenic Nicotiana tabacum (tobacco) BrightYellow-2 (BY-2) cells which ectopically expressed GmMATE1-YFP or GmMATE2-YFP. FMTM4-64 was used to visualize the plasma membrane and vacuolar membrane [2]. After 24 h of incubation, FMTM4-64 signals appeared in the vacuolar membrane and co-localized with the signals from GmMATE1-YFP and GmMATE2-YFP for both the C08 and W05 allelic forms (Figure 3).

Figure 3. Subcellular localizations of GmMATE1-YFP and GmMATE2-YFP were determined using confocal microscopy. The cells were incubated with FMTM4-64 in the dark at room temperature for 24 h before imaging. DIC, differential interference contrast. Signals from GmMATE-YFP and FMTM4-64 were illustrated in green and magenta, respectively.
Based on the results of yeast uptake assays and subcellular localization studies, the allelic differences between W05 and C08 did not seem to result in the differential accumulation of isoflavones between GmMATEW05 and GmMATEC08 expressing yeasts. Therefore, only the C08 allelic form was employed in subsequent functional analyses.
3. GmMATE1 and GmMATE2 Were Expressed in Developing Pods, Seeds, and the Seed Coat
The expression of GmMATE1 and GmMATE2 in pods, seeds, and the seed coat at different developmental stages were quantified using RT-qPCR. Although GmMATE1 and GmMATE2 showed different expression patterns among pod, seed, and seed coat during the development; in general, the expression of GmMATE1 and GmMATE2 could be found in all three types of tissues and the expression tended to diminish 40 days after flowering (Figure 4).
The tissue-specific expression of GmMATE1C08 was studied using transgenic soybean expressing β-glucuronidase (GUS) driven by the native promoter of GmMATE1C08 (GmMATE1C08pro-GUS). The GUS stain could be observed clearer in soybean pods around the main vein, compared to seeds and the seed coat (Figure 4). As an additional confirmation, the same construct was expressed in transgenic A. thaliana. The GUS stain was found mainly in the funicle of siliques (the equivalents of soybean pods in A. thaliana [3] (Figure 4). The funicle is the compartment through which the secondary metabolites are transported into developing seeds[4] .
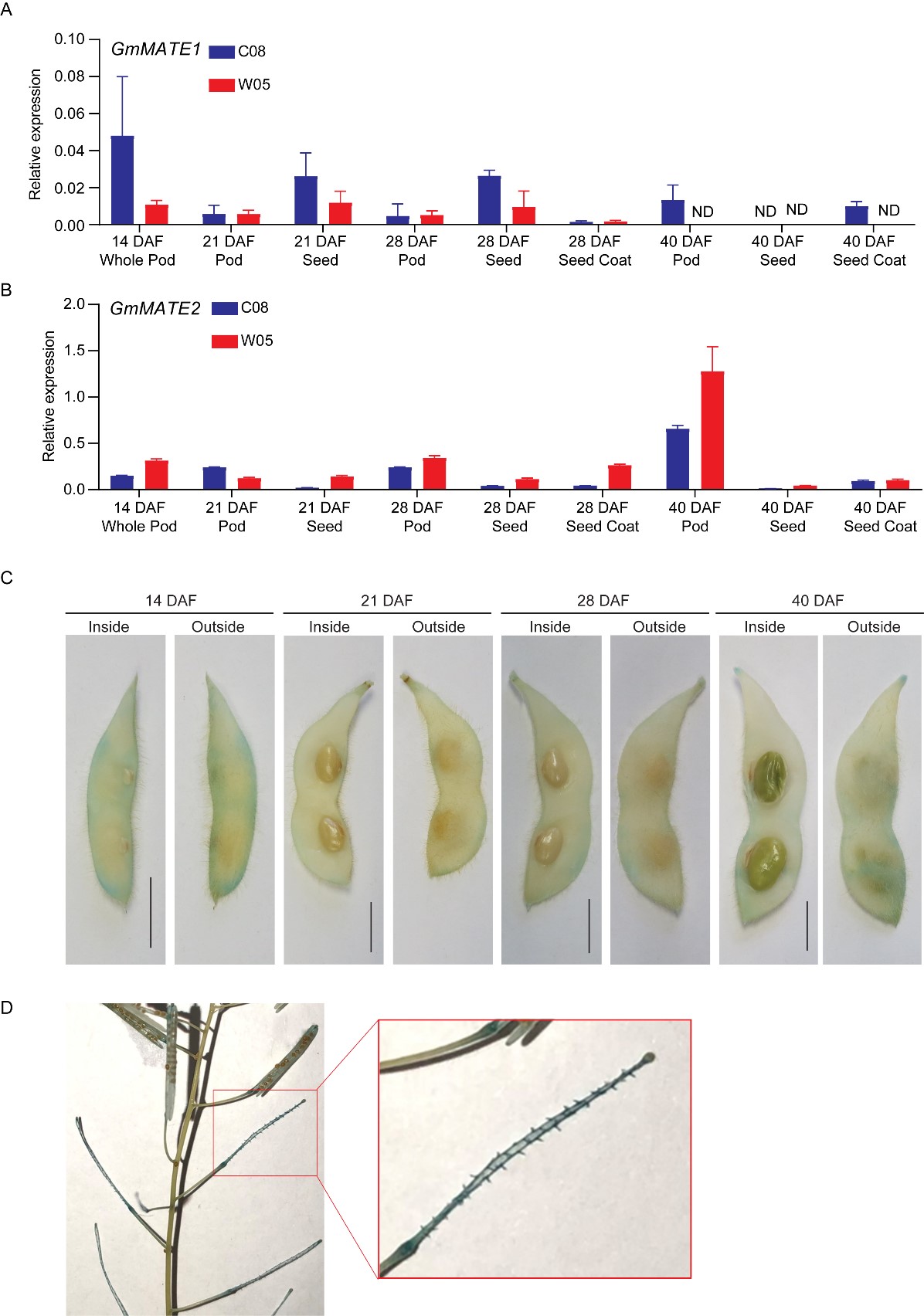
Figure 4. Relative expressions of GmMATE1 and GmMATE2 in different tissues within the developing soybean seed pod. The relative expression of (A) GmMATE1 and (B) GmMATE2 in different tissues of the developing pod, normalized to the reference gene, VPS, using the 2−ΔCt method. DAF, days after flowering. Values shown are the averages of three technical replicates ± SEM. Similar expression trends were obtained in another biological repeat. (C,D) Photos showing the GUS staining of (C) transgenic soybean pods and (D) transgenic A. thaliana expressing β-glucuronidase (GUS) driven by the native GmMATE1C08 promoter. Similar results were obtained from two independent transgenic lines. Scale bar = 1 cm.
4. Manipulation of GmMATE1 in Soybean Significantly Altered the Isoflavone Contents in Seeds
The native GmMATE1 gene was knocked out using the CRISPR/Cas9 system or overexpressed under the control of a constitutive 35S promoter. A guide RNA, targeting the first exon of GmMATE1, was used to generate knockout mutants in the soybean germplasm, Williams 82 (W82). After screening, mate1 knockout lines were generated in the W82 background (Figure 5). The allelic form of GmMATE1C08 is the same as that of W82. Significantly lower total isoflavone contents were detected in the knockout mutant compared to the wild type (Figure 5). In the GmMATE1C08 over-expressor (GmMATE1C08OX), statistically significant enhancement in total isoflavones (genistein, daidzein, glycitein, genistin, daidzin, glycitin, malonylgenistin, malonyldaidzin, and malonylglycitin) was observed (Figure 5), suggesting an overall improvement in isoflavone accumulation.
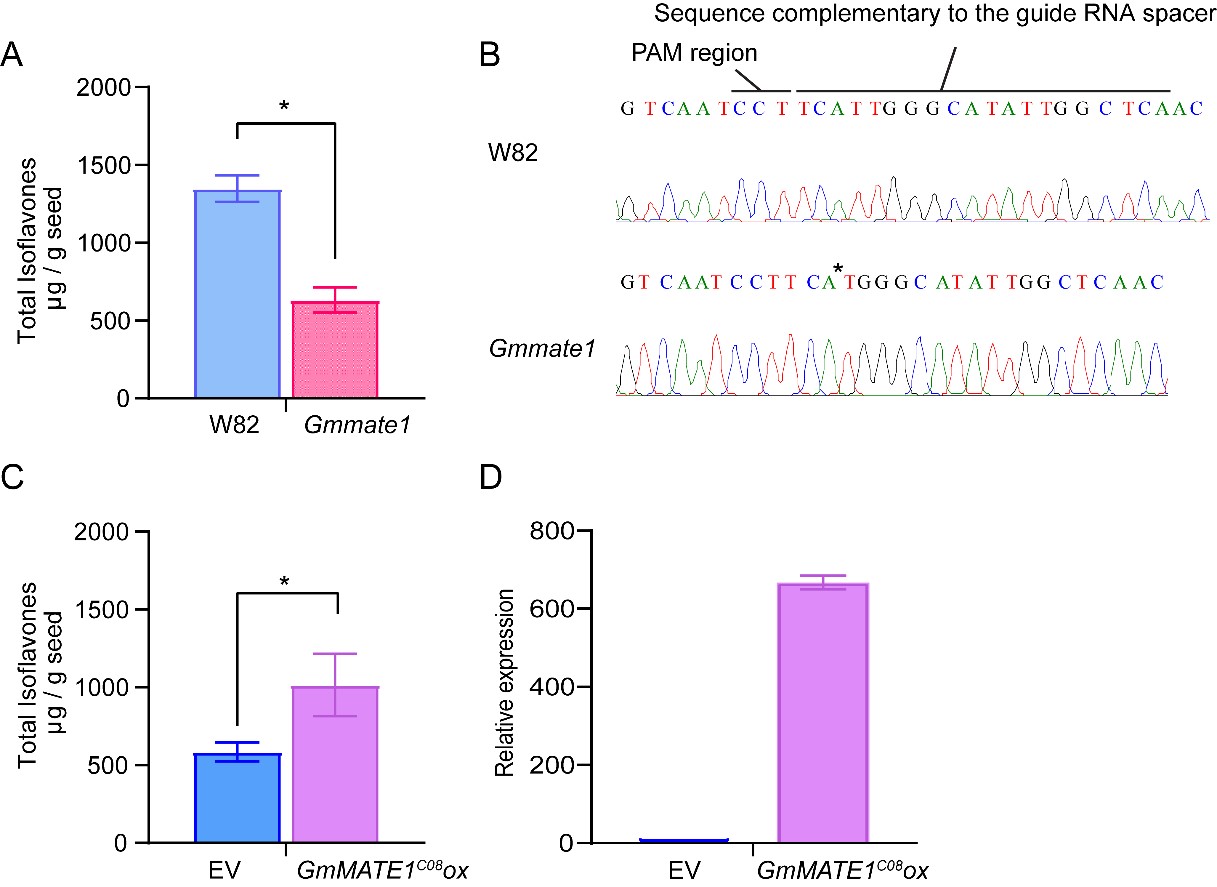
Figure 5. GmMATE1 is responsible for determining seed isoflavone contents. (A) The total isoflavone contents in the CRISPR/Cas9-edited GmMATE1 mutant was significantly lower than those in its wild-type, W82. Values shown were the means of at least three independently collected batches of seeds ± SEM. Significant differences were determined by Mann-Whitney test. *, p < 0.05. (B) The mutation site in the GmMATE1C08 gene, with the single-nucleotide deletion marked with an asterisk. (C) The levels of isoflavones in the transgenic soybean overexpressing GmMATE1C08 were significantly higher than those in the empty vector control, both in the W82 background. Values shown were the means of four independently collected batches of seeds ± SEM. Significant differences were determined by Mann-Whitney test. *, p < 0.05. (D) Relative expression levels of GmMATE1 in the GmMATE1C08-overexpressor and the empty vector control in the W82 background. The expression levels were normalized to the reference gene, VPS, using the 2−ΔΔCt method.
The above findings are published in [5].
References
- Li, M. W.; Munoz, N. B.; Wong, C. F.; Wong, F. L.; Wong, K. S.; Wong, J. W. H.; Qi, X. P.; Li, K. P.; Ng, M. S.; Lam, H. M. QTLs regulating the contents of antioxidants, phenolics, and flavonoids in soybean seeds share a common genomic region. Front. Plant Sci. 2016, 7, 854.
- Bolte, S.; Talbot, C.; Boutte, Y.; Catrice, O.; Read, N. D.; Satiat-Jeunemaitre, B. FM-dyes as experimental probes for dissecting vesicle trafficking in living plant cells. J. Microsc. 2004, 214, 159-173.
- Christiansen, L. C.; Dal Degan, F.; Ulvskov, P.; Borkhardt, B. Examination of the dehiscence zone in soybean pods and isolation of a dehiscence-related endopolygalacturonase gene. Plant Cell Environ. 2002, 25, 479-490.
- Xie, H.; Wang, D.; Qin, Y.; Ma, A.; Fu, J.; Qin, Y.; Hu, G.; Zhao, J. Genome-wide identification and expression analysis of SWEET gene family in Litchi chinensis reveal the involvement of LcSWEET2a/3b in early seed development. BMC Plant Biol. 2019, 19, 499.
- Ng, M.S.; Ku, Y.S.; Yung, W.S.; Cheng, S.S.; Man, C.K.; Yang, L.; Song, S.; Chung, G.; Lam, H.M. MATE-type proteins are responsible for isoflavone transportation and accumulation in soybean seeds. Int. J. Mol. Sci. 2021, 22, 12017.
