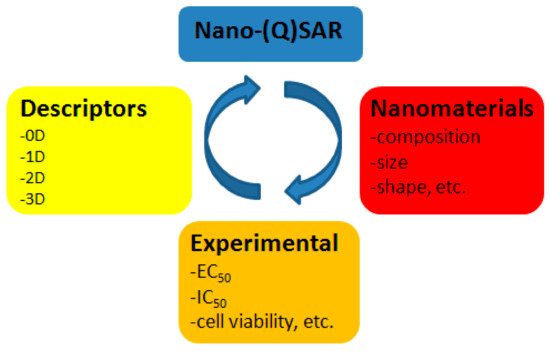
Nanomaterials and nanoparticles (NPs) possess unique physico-chemical properties (size, shape, chemical composition, physiochemical stability, crystal structure, surface area, surface energy, and surface roughness), which give them beneficial characteristics. Quantitative structure-activity relationship, or QSAR, is an area of molecular modeling that studies relationships between structure and activity using mathematical statistics and machine learning methods. QSAR is efficiently used to predict toxicity of chemical substances.
- engineered nanomaterials
- safety of nanomaterials
- toxicological tests
- descriptors
- QSAR
- machine learning
- modeling
1. Introduction
2. Metal Oxides
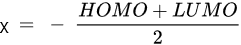
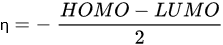
|
Source |
Dataset |
Endpoint of Cytotoxicity Measurement |
n |
|---|
|
Source |
Dataset | R |
Cell Type | 2 1 |
Source Endpoint of Cytotoxicity Measurement |
Dataset n |
Software |
Statistical Method |
Descriptors |
|---|
|
Source |
Dataset | Cell Type |
Cell Type Endpoint of Cytotoxicity Measurement |
Endpoint of Cytotoxicity Measurement n |
R2 |
Software |
n |
Software 2 |
R2 |
R2 |
Statistical Method |
Descriptors |
|---|
Software | Software | Statistical Method |
Statistical Method Descriptors |
|||||
|---|---|---|---|---|---|---|---|---|
Descriptors | ||||||||
Statistical Method | Descriptors |
|
Escherichia coli |
|
[69 |
Monocytes, hepatocytes, endothelial, and smooth muscle cells |
Cellular viability |
51 |
0.72 |
WinSVM, ISIDA |
][74] |
[ | |||||||||||||
|
Salmonella typhimurium TA100 |
Salmonella typhimurium TA100 Reverse mutation test TA100 |
Reverse mutation test TA100 24 |
Human embryonic kidney cells HEK293 |
Cell viability (%) 44 0.65–0.81 |
0.60–0.78 |
40 CORAL |
SVM classification and k Nearest Neighbors (kNN) regression |
Monte Carlo |
0.80–0.93 CORAL |
CORAL Monte Carlo Size, zeta potential, R1 and R2 relaxivities |
Quasi-SMILES |
|||||||||
Quasi-SMILES | ||||||||||||||||||||
Monte Carlo | Quasi-SMILES | |||||||||||||||||||
LD50 |
PaCa2 human pancreatic cancer cells, U937 macrophage cell lines, primary human macrophages, HUVEC human umbilical vein endothelial cells |
7 |
][76 |
0.979 |
] |
Salmonella typhimurium TA100 |
- |
Multiple linear regression (MLR) |
Metal cation charge |
|||||||||||
Reverse mutation test TA100 |
30 |
0.53–0.64 |
CORAL |
Monte Carlo |
Quasi-SMILES |
|||||||||||||||
|
S. typhimurium TA100 |
Cellular uptake |
Reverse mutation test TA100 109 |
20 0.65–0.80 |
0.76 |
CORAL |
Monte Carlo |
||||||||||||||
WinSVM, ISIDA | Quasi-SMILES |
Human kidney cells HK-2 | ||||||||||||||||||
Cell viability (%) | SVM classification and k Nearest Neighbors (kNN) regression |
42 |
0.83–0.89 |
CORAL |
Monte Carlo Lipophilicity, number of double bonds |
Quasi-SMILES |
[20] |
|||||||||||||
|
[20] |
[56 [ |
LD50 |
]72] |
17 |
[[74 |
0.862 |
60][ |
MATLAB |
] |
MLR |
Enthalpy of formation of a gaseous cation |
|||||||||
Smooth muscle cells |
79] Cell apoptosis |
31 |
0.81 |
[86 Salmonella typhimurium TA100 |
S. typhimurium TA100 |
] |
16HBE, A549, HaCaT, NRK-52E, and THP-1 Reverse mutation test TA100 - |
Reverse mutation test TA100 |
EC25 44 |
20 0.60–0.78 MLR and Bayesian regularized artificial neural network |
CORAL |
19 0.63–0.76 |
CORAL Monte Carlo IFe2O3 |
Quasi-SMILES |
||||||
Monte Carlo | , I | dextran |
Quasi-SMILES | |||||||||||||||||
0.83 | CORAL | , and I | surf.chg |
Monte Carlo |
Quasi-SMILES |
|||||||||||||||
] [75 |
||||||||||||||||||||
|
][80] |
LD50 |
Monocytes, hepatocytes, endothelial, and smooth muscle cells |
17 |
Four types of normal human lung cells (BEAS-2B, 16HBE14o-, WI-38, and HBE) |
0.741–0.838 |
Cell viability (%) |
E. coli WP2 uvrA/pKM101 |
[82] |
CORAL |
Monte Carlo |
SMILES-based optimal descriptor |
|||||||||
[ | 86] | Cellular viability |
44 |
276 |
16HBE, A549, HaCaT, NRK-52E, and Reverse mutation test WP2 uvrA/pKM101 - |
0.60–0.80 |
20 - |
CORAL |
0.68–0.82 |
CORALNaive Bayesian classifier |
Monte Carlo |
Monte Carlo Primary size, spin-lattice and spin-spin relaxivities, zeta potential |
Quasi-SMILES |
Quasi-SMILES |
||||||
|
[20] |
6. Silica Nanomaterials
THP-1 | |||||||||||||||||||
EC | 25 |
19 |
0.87 |
R |
RF |
Aspect ratio and zeta potential |
|||||||||||||
[60][ | |||||||||||||||||||
64] |
[LD50 |
17 |
] |
0.933 |
Zebrafish embryo |
Human embryonic kidney cell line (HEK293) |
Minitab 16 |
24 h post-fertilization mortality |
Cell viability (%) 82 |
- | MLR |
ABMiner |
Energy gap, hardness, softness, electronegativity, and electrophilicity index |
||||||
40 | Numerical prediction |
0.80–0.95 | Concentration, shell composition, surface functional groups, purity, core structure, and surface charge |
CORAL |
Monte Carlo |
Quasi-SMILES |
|||||||||||||
|
[20] |
LD50 |
17 |
0.81–0.90 |
Mammalian cell lines |
- |
- |
MLR |
STATISTICA v.6 |
Electronegativity, charge of the metal cation corresponding to a given oxide |
||||||||||
TC | 50 | LDA |
Molar volume, polarizability, and size of the particles |
||||||||||||||||
|
[20] |
] |
LD50 |
17 |
Algae, bacteria, cell lines, crustaceans, plants, fish, and others |
CC50, EC50, IC50 |
0.93 |
, TC50, LC50 |
RandomForest package |
Random forest (RF) |
S |
36488 |
- |
STATISTICA | 1—unbonded two-atomic fragments [Me] … [Me], which were encoded based on Simplex representation of molecular structures (SiRMS)-derived descriptors [33][34][34,35], describing distance where potential reaches minimum at van der Waals interactions; rw—Wigner–Seitz radius; ρ—mass density; (CPP)—cation polarizing power; S2—SiRMS-derived electronegativity aligned descriptor of oxides molecules—in a sense of the acid-base property of oxides (this parameter increases with a number of oxygens in molecule); S3—tri-atomic fragments [Me]-[O]-[Me], which were encoded by SiRMS-derived descriptors, encoding electronegativity; and (SV)—proportion of surface molecules to molecules in volume |
|||||
LDA | Molar volume, polarizability, size of NPs, electronegativity, hydrophobicity, and polar surface area of surface coating |
[20] |
LD50 |
17 |
0.955 |
Ensemble learning |
Oxygen percent, molar refractivity, and polar surface area |
||||||||||||
|
Bacteria, algae, crustaceans, fish, and others |
EC50, IC50, TC50, LC50 |
5520 |
- |
STATISTICA |
LDA |
Molar volume, electronegativity, polarizability, and nanoparticle size |
[20] |
LD50 |
17 |
- |
MATLAB |
Read-across |
Ionization enthalpy of the detached metal atoms |
||||||
|
[18] |
[20] |
LD50 |
17 | ||||||||||||||||
|
Algae, bacteria, fungi, mammal cell lines, crustaceans, plants, fishes, and others |
CC50, EC50, IC50, TC50, LC50 |
54371 |
- |
STATISTICA |
Artificial neural network |
Polar surface area, hydrophobicity, atomic weight, atomic van der Waals radius, electronegativity, and polarizability |
|||||||||||||
|
[65][ | 0.889–0.982 | 69] |
CORAL |
Danio rerio, Daphnia magna, Pseudokirchneriella subcapitata, and Staphylococcus aureus |
LC50, EC50, MIC (minimum inhibitory concentration) |
MLR |
SMILES-based optimal descriptor |
||||||||||||
400 | - |
Weka |
Functional tree, C4.5 decision tree, random tree, and CART |
Molecular polarizability, accessible surface area, and solubility |
[20] |
LD50 |
16 |
0.91 |
- |
MLR |
Enthalpy of formation of a gaseous cation (ΔH | ||||||||
|
E. coli and Chinese hamster ovary (CHO-K1) cells |
EC50, MIC | Me+ | ), charge of the metal cation (χ |
17 | ox), and pEC50 of HaCaT |
||||||||||||||
0.94 | R | Nonlinear least-squaress |
Size and specific surface area (Brunauer-Emmett-Teller surface) |
[20] |
LD50 |
16 |
0.879 |
SYBYL X1.1 and SPSS statistics v.17 |
MLR |
Enthalpy of formation of a gaseous cation (ΔHme+) and polarization force (Z/r) |
|||||||||
|
[20] |
LD50 |
16 |
0.79 |
CORAL |
Monte Carlo |
Quasi-SMILES |
|||||||||||||
|
[20] |
LD50 |
17 |
0.92 |
- |
Counter propagation artificial neural network |
Metal electronegativity by Pauling scale, number of metal atoms in oxide, number of oxygen atoms in oxide, and charge of metal cation |
|||||||||||||
|
[20] |
LD50 |
17 |
0.968 |
- |
RF |
Oxygen in weight percentage and enthalpy of formation of a gaseous cation |
|||||||||||||
|
[20] |
LD50 |
17 |
0.877 and 0.903 |
- |
MLR and support vector machines (SVM) |
HOMO energy, α-LUMO and β-LUMO energy, the average of α-LUMO and β-LUMO, the energy gap between the frontier molecular orbitals ∆E, and molar heat capacity |
|||||||||||||
|
[8] |
[20] |
LD50 |
17 |
0.93 |
- |
Partial least squares (PLS) |
Charge of metal ion, metal ion charge-based SiRMS, number of oxygen atoms in brutto formula weighted by ionic potential, covalent index weighted by charge of metal ion, molecular weight of metal oxide weighed by size of nanoparticle, squared thickness of interfacial layer, van der Waals repulsion weighted by size of nanoparticle, and Wigner-Seitz radius weighted by size of nanoparticle |
||||||||||||
|
LD50 |
17 |
0.87 |
Self-written program |
MLR |
Electronegativity of metal and electronegativity of metal oxide |
||||||||||||||
|
IC50 |
24 |
- |
R |
SVM |
Conduction band energy and hydration enthalpy (ΔHhyd) |
||||||||||||||
|
Human keratinocyte cell line (HaCaT) |
|||||||||||||||||||
|
LD50 |
18 |
0.96 |
RandomForest package |
RF |
S1, rw, ρ, (CI)—covalent index of the metal ion, S2, and (AP)—aggregation parameter |
||||||||||||||
|
LD50 |
18 |
- |
MATLAB |
Read-across |
Mulliken’s electronegativity |
||||||||||||||
|
LD50 |
18 |
0.93 |
- |
MLR |
Enthalpy of formation of metal oxide, Mulliken’s electronegativity |
||||||||||||||
|
[18] |
LD50 |
18 |
0.961–0.999 |
CORAL |
MLR |
SMILES-based optimal descriptor |
|||||||||||||
|
LD50 |
16 |
0.88 |
- |
MLR |
Enthalpy of formation of metal oxide (ΔHf) nano-cluster, electronic chemical potential of the cluster, and pEC50 of E. coli |
||||||||||||||
|
LD50 |
16 |
0.79 |
CORAL |
Monte Carlo |
Quasi-SMILES |
||||||||||||||
|
LD50 |
18 |
0.918 |
- |
RF |
10-based logarithm of solubility measured in mol/L (LogS), topological polar surface area (TPSA), Mulliken’s electronegativity |
||||||||||||||
|
[8] |
LD50 |
18 |
0.83 |
- |
PLS |
Atom charge-based SiRMS descriptor, charge of the atom weighted by the bond ionicity, charge of metal ion weighted by ionicity of bond, squared ionic potential, ion change-based SiRMS descriptor, number of oxygen atoms in brutto formula per interfacial layer, mass density weighted by ionicity of bond, Wigner-Seitz radius weighted by ionicity of bond, and ionicity of bond based SiRMS |
|||||||||||||
|
Cell viability (%) |
21 |
- |
CORAL |
Hierarchical cluster analysis (HCA) and min–max normalization |
Quasi-SMILES |
||||||||||||||
|
Transformed bronchial epithelial cells (BEAS-2B) |
|||||||||||||||||||
|
% of membrane-damaged cells |
9 |
- |
Weka |
RF |
Atomization energy of the metal oxide, period of the nanoparticle metal, nanoparticle primary size, and nanoparticle volume fraction |
||||||||||||||
|
[6] |
[6] |
Cell viability (%) |
24 |
- |
- |
Regression tree |
Metal solubility and energy of conduction |
||||||||||||
|
[6] |
Cell viability (%) |
24 |
- |
RandomForest package |
RF |
Mass density, covalent index, cation polarizing power, Wigner–Seitz radius, surface area-to-volume ratio, aggregation parameter, and tri-atomic descriptor of atomic charges |
|||||||||||||
|
LD50 |
24 |
- |
RapidMiner |
SVM |
Conduction band energy and ionic index of metal cation |
||||||||||||||
|
% of membrane-damaged cells |
24 |
0.68 |
CORAL |
Monte Carlo |
SMILES-based optimal descriptor, dose, and exposure time |
||||||||||||||
|
Cell viability (%) |
21 |
0.713–0.733 |
CORAL |
HCA and min-max normalization |
Quasi-SMILES |
||||||||||||||
|
Murine myeloid cells (RAW 264.7) |
|||||||||||||||||||
|
[6] |
[6] |
Cell viability (%) |
24 |
- |
- |
Regression tree | |||||||||||||
1681 | Metal solubility and energy of conduction |
[6] |
Cell viability (%) |
24 |
- |
RandomForest package |
RF |
Mass density, molecular weight, aligned electronegativity, covalent index, surface area, surface area-to-volume ratio, two-atomic descriptor of van der Waals interactions, tetra-atomic descriptor of atomic charges, and size in DMEM |
|||||||||||
|
LD50 |
24 |
- |
RapidMiner |
SVM |
Conduction band energy and ionic index of metal cation |
||||||||||||||
|
Lactate dehydrogenase (LDH) release |
25 |
- |
R |
PLS |
Metal cation charge, hydration rate, radius of the metallic cation, and Pauling electronegativity |
||||||||||||||
|
Rat L2 lung epithelial cells and rat lung alveolar macrophages |
|||||||||||||||||||
|
Membrane damage (units L−1) |
42 |
- |
- |
Multivariate linear regression and linear discriminant analysis (LDA) |
Size, concentration, size in phosphate buffered saline, size in water, and zeta potential |
||||||||||||||
|
Membrane damage (units L−1) |
42 |
- |
- |
MLR and simple classification |
Size, concentration, size in phosphate buffered saline, and size in water |
||||||||||||||
1 Missing R2 value means that an SAR model was built instead of QSAR. 2 If software record is missing, then it was not mentioned in the original paper.
3. Other Metal-Containing Nanoparticles
4. Multi-Walled Carbon Nanotubes (MWCNTs)
5. Fullerenes
|
Source |
Dataset |
Cell Type |
Endpoint of Cytotoxicity Measurement |
n |
R2 |
|---|
