Flavonoids have long been a major focus of research into secondary metabolism. We present a systematic summary of what is known of the flavonoid biosynthetic pathway in plants, presenting a model of flavonoid biosynthesis that includes eight branches (stilbene, aurone, flavone, isoflavone, flavonol, phlobaphene, proanthocyanidin, and anthocyanin biosynthesis) and four important intermediate metabolites (chalcone, flavanone, dihydroflavonol, and leucoanthocyanidin).
- flavonoids
- biosynthesis
- molecular structure
- biosynthetic enzyme
- gene regulation
1. Introduction
- Introduction
Flavonoids comprise a group of phenylpropanoids that as water-soluble pigments stored in the vacuoles of plant cells [1]. Except for the stilbenes (a class of flavonoids), which has a C6-C2-C6 structure (Figure 1), the basic structure of flavonoids consists of a C6-C3-C6 carbon skeleton (Figure 1) comprising two 6-carbon benzene rings (rings A and B) linked by a 3-carbon heterocyclic ring (ring C) [2]. Flavonoids can be classified into 12 subgroups—chalcones, stilbenes, aurones, flavanones, flavones, isoflavones, phlobaphenes, dihydroflavonols, flavonols, leucoanthocyanidins, proanthocyanidins, and anthocyanins (Figure 1) [3, [3][4]4]—based on the degree of oxidation of the heterocyclic ring and the number of hydroxyl or methyl groups on the benzene ring. At the same time, various modifications (glycosylation, acylation, and others) and molecular polymerization lead to the formation of a large number of flavonoid compounds [5][6][5, 6]. To date, more than 9,000 plant flavonoids have been isolated and identified [7].
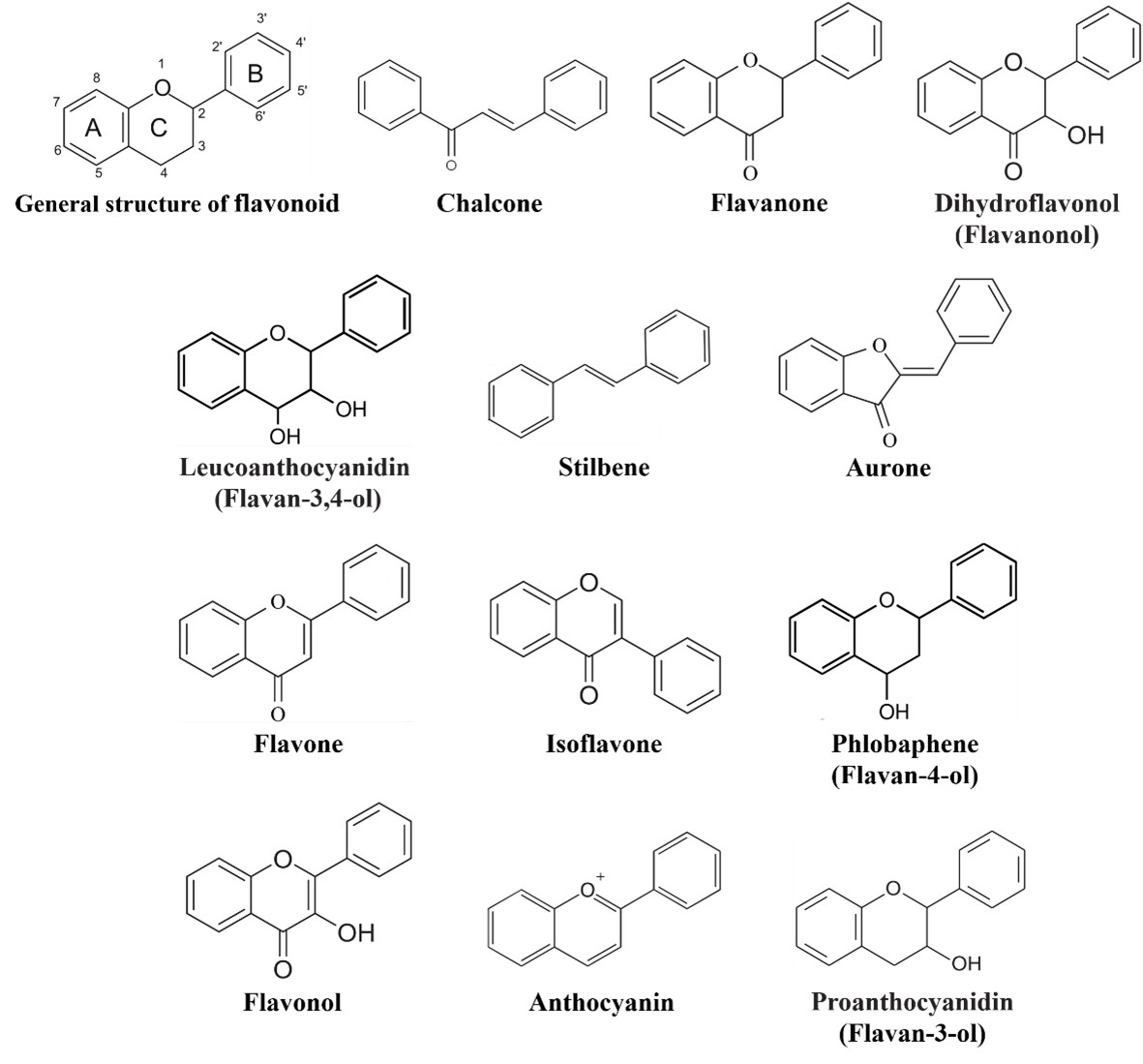
Figure 1. General structure of flavonoids
Some flavonoids play an important role in plant development and defense. Flavonoids constitute one of the main pigments in plants, such as the anthocyanins (red, orange, blue, and purple pigments); chalcones and aurones (yellow pigments); and flavonols and flavones (white and pale-yellow pigments), that impart on plants a wide variety of colors [8] [8]. Flavonoids, as phytoalexins or antioxidants, have reactive oxygen species (ROS) scavenging ability [9] and protect plants against damage from biotic and abiotic stresses, including UV irradiation, cold stress, pathogen infection, and insect feeding [10][11][12][10- 12]. In plants, flavonoids can also act as signaling molecules, attracting insects for pollination and participating in auxin metabolism [13]. Plant flavonoids also have widespread use in daily life, such as for food and medicinal purposes. For instance, anthocyanins and proanthocyanidins are important edible pigments and taste-regulating components in food and wine [4], while plant flavonoids, administered as active ingredients, can help delay the aging of the nervous system, immune organs, reproductive system, liver, and skin, and also contribute to the prevention of osteoporosis, cardiovascular disease, Alzheimer’s disease, and breast cancer [14][15][16][14- 16].
2. Flavonoid biosynthesis in plants
- Flavonoid biosynthesis in plants
Flavonoids have long been a major focus of research into secondary metabolism. On PubMed, performing a search using 'flavonoid' as a search term retrieves more than 10,000 articles in both 2019 and 2020. Recent decades have witnessed a considerable renewed interest in flavonoid biosynthesis in plants. In this review, we present a systematic summary of what is known of the flavonoid biosynthetic pathway in plants, presenting a model of flavonoid biosynthesis that includes eight branches and four intermediate metabolites (Figure 2), thereby providing a theoretical basis for the genetic improvement of flavonoid metabolism as well as improving our understanding of their functions and potential uses.
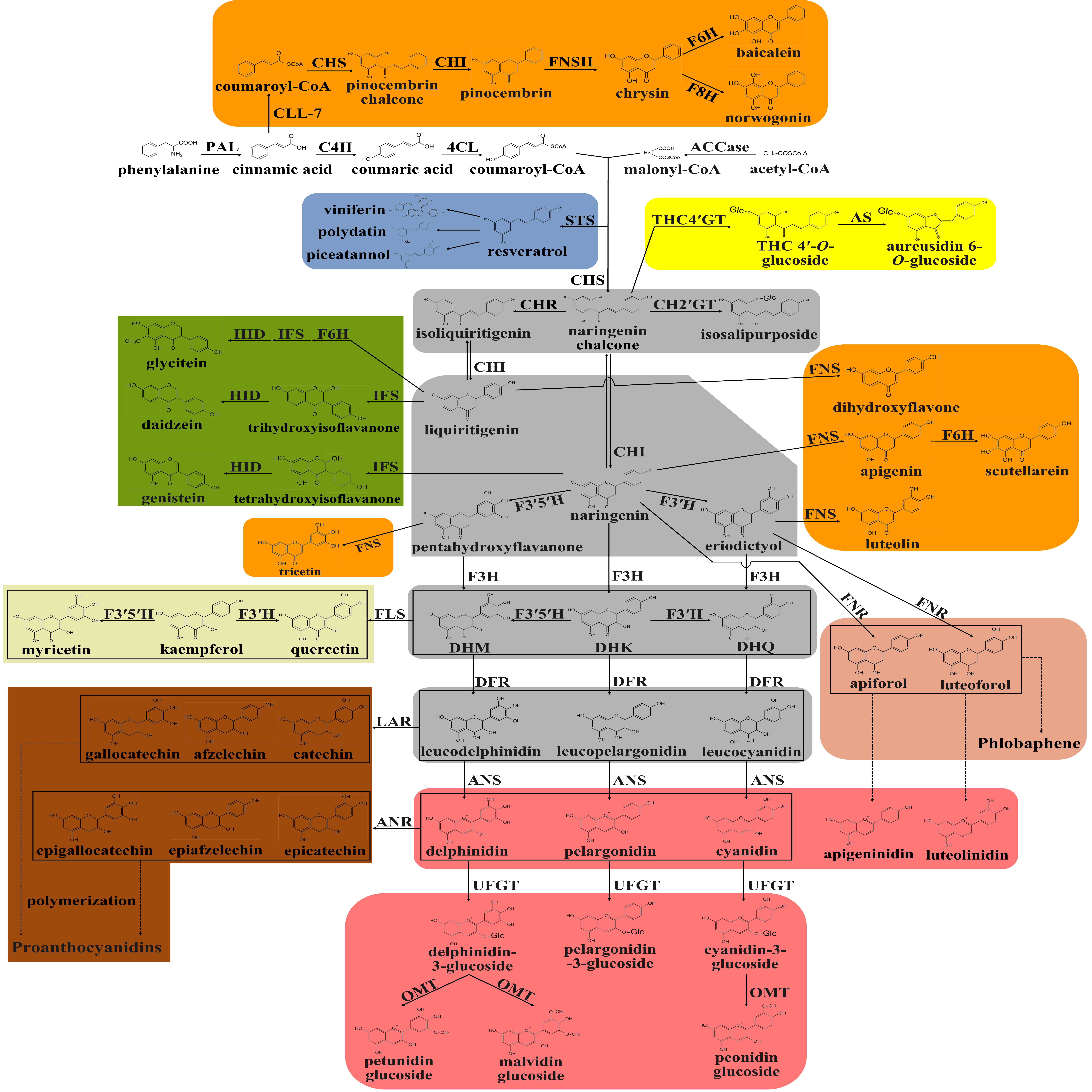
Figure 2. The flavonoid biosynthetic pathway in plants contains eight branches (represented by the eight colored boxes) and four important intermediate metabolites (gray boxes). The enzyme names and flavonoid compounds are abbreviated as follows: PAL, phenylalanine ammonia lyase; C4H, cinnamic acid 4-hydroxylase; 4CL, 4-coumarate: CoA ligase; ACCase, acetyl-CoA carboxylase; STS, stilbene synthase; CHS, chalcone synthase; CHR, chalcone reductase; CH2′GT, chalcone 2′-glucosyltransferase; CH4′GT, chalcone 4′-O-glucosyltransferase; AS, aureusidin synthase; CHI, chalcone isomerase; FNS, flavone synthase; CLL-7, cinnamate–CoA ligase; F6H, flavonoid 6-hydroxylase; F8H, flavonoid 8-hydroxylase; IFS, isoflavone synthase; HID, 2-hydroxyisoflavanone dehydratase; FNR, flavanone 4-reductase; F3H, flavanone 3-hydroxylase; F3′5′H, flavanone 3′,5′-hydroxylase; DHK, dihydrokaempferol; DHQ, dihydroquercetin; DHM, dihydromyricetin; FLS, flavonol synthase; DFR, dihydroflavonol 4-reductase; ANS, anthocyanidin synthase; UFGT, UDP-glucose flavonoid 3-O-glucosyltransferase; OMT, O-methyl transferases; LAR, leucoanthocyanidin reductase; ANR, anthocyanidin reductase.
3. Transcriptional regulation of flavonoid biosynthesis in plants
- Transcriptional regulation of flavonoid biosynthesis in plants
Transcriptional control plays a central role in the modulation of flavonoid biosynthesis (Figure 3). The MBW complex, composed of MYB, bHLH, and WD40, is the main transcriptional regulator in flavonoid biosynthesis [17] [17]. MYB transcription factors have a conserved MYB domain in the N-terminus that is required for DNA binding and interaction with other proteins [18]. Members of the R2R3-MYB group are mainly involved in regulating flavonoid metabolism [19][19]. The overexpression of AN4 (a R2R3-MYB-encoding gene) can enhance anthocyanin biosynthesis by promoting the expression of anthocyanin biosynthesis genes, such as CHS, CHI, F3H, and DFR [20]. In Cucumis sativus, the R2R3-MYB transcription factor CsMYB60 induced the expression of CsFLS and CsLAR by binding to their promoters, thereby promoting flavonol and proanthocyanidin biosynthesis [21]. MYB transcription factors also act as repressors in the regulation of flavonoid biosynthesis. For instance, in the apple (Malus domestica), MdMYB15L was reported to interact with MdbHLH33 and inhibit the promotion of the MdbHLH33-MYB-WD40 (MBW) complex, thereby also suppressing anthocyanin biosynthesis [22].
bHLH transcription factors have been shown to participate in the regulation of flavonoid biosynthesis. The transient expression of DhbHLH1 induces anthocyanin synthesis in the white petals of Dendrobium hybrids [23]. In Dianthus caryophyllus, meanwhile, the “red speckles and stripes on white petals” phenotype results from the local expression of bHLH, which promotes the expression of DFR and that of downstream enzymes in the anthocyanin biosynthetic pathway [24].
WD40, widely present in eukaryotic cells, contains multiple tandem repeats of a WD motif and interacts with other proteins through its WD domain [1]. Generally, WD40 does not directly bind to target gene promoters, forming instead a complex with MYB and bHLH in the regulation of flavonoid biosynthesis. The WD40 protein TTG1 regulated anthocyanin metabolism through MYB/bHLH/TTG1 complex [25]. Moreover, in tomato, the WD40 protein SlAN11 was shown to induce anthocyanin and proanthocyanidin biosynthesis and limit flavonol accumulation by repressing FLS expression [26].
Also in tomato, besides the MBW complex, the transcription factors NF-YA, NF-YB, and NF-YC can reportedly form a NF-Y protein complex that binds to the promoter of the CHS1 gene, thereby regulating flavonoid synthesis and affecting tomato peel color [27]. Additionally, the ethylene response factors Pp4ERF24 and Pp12ERF96, through interacting with PpMYB114, potentiated the PpMYB114-mediated accumulation of anthocyanin in pear [28]. In the tea plant, UV-B irradiation-mediated bZIP1 upregulation leads to the promotion of flavonol biosynthesis by binding to the promoters of MYB12, FLS, and UGT and activating their expression; under shading, meanwhile, PIF3 inhibited flavonol accumulation by activating the expression of MYB7, which encodes a transcriptional repressor [29]. In peach, NAC1 was shown to regulate anthocyanin pigmentation through activating the transcription of MYB10.1, while NAC1 was repressed by SPL1 [30]. In the pear, PyWRKY26 interacts with PybHLH3 and activates the expression of PyMYB114, resulting in anthocyanin biosynthesis [31]. The BTB/TAZ protein MdBT2 represses anthocyanin biosynthesis, and MdGRF11 interacts with, and negatively regulates, MdBT2, leading to an increase in the expression of anthocyanin biosynthesis-related genes via the enhancement of the abundance of MdMYB1 protein [32]. SlBBX20 can bind the SlDFR promoter and directly activate its expression, which augments anthocyanin biosynthesis, while SlCSN5, a subunit of the COP9 signalosome, induces the degradation of SlBBX20 by enhancing its ubiquitination [33]. MdARF19 modulates anthocyanin biosynthesis by binding to the promoter of MdLOB52 and further activating its expression [34]. BES1, a positive regulator in brassinosteroid signaling, inhibits the transcription of the MYB proteins MYB11, MYB12, and MYB111, thereby decreasing flavonol biosynthesis [35].[35]
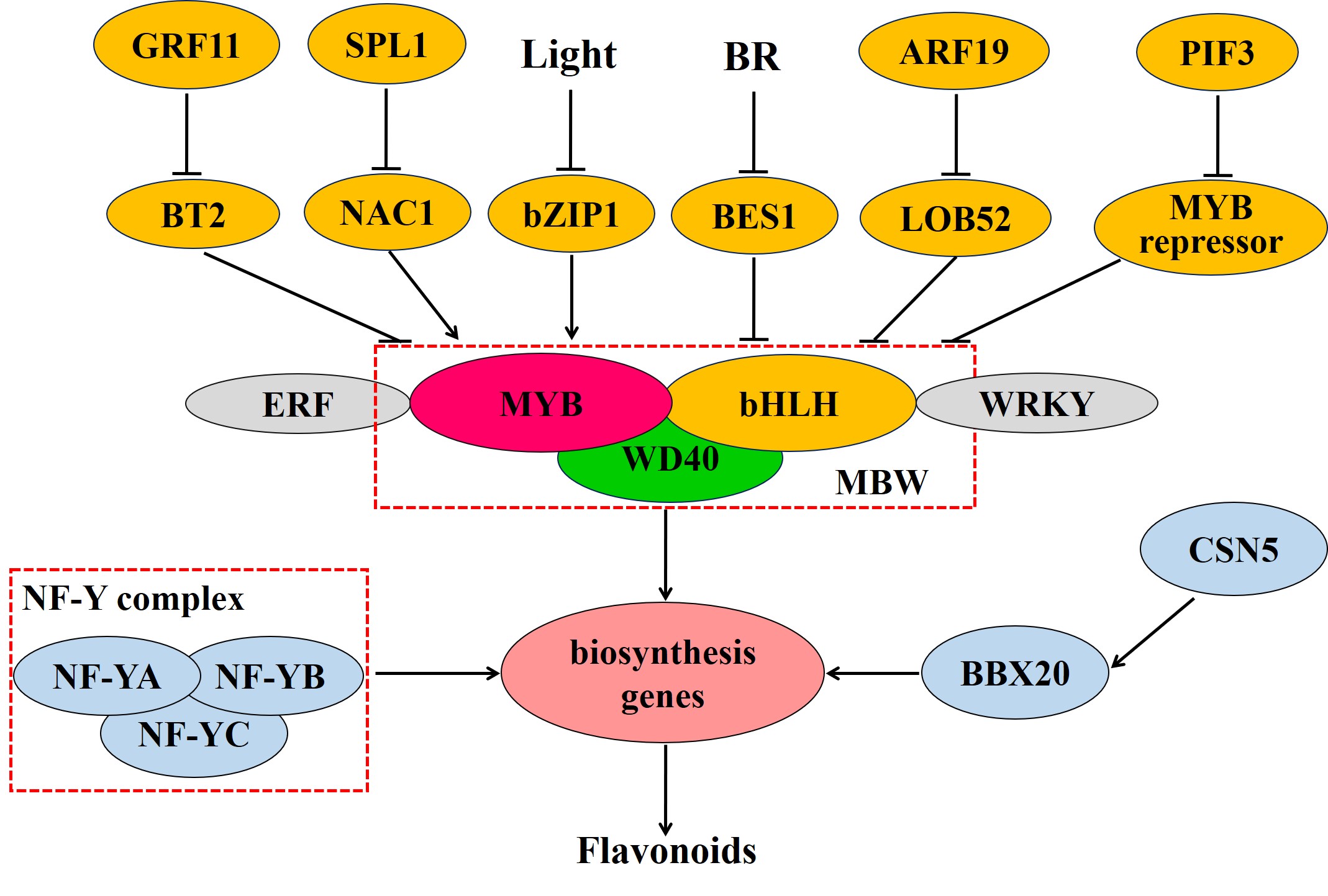 Figure 3. Transcriptional regulation of flavonoid biosynthesis in plants. Abbreviations are as follows: MYB, v-myb avian myeloblastosis viral oncogene homolog; bHLH, basic helix-loop-helix; NF-Y, nuclear factor Y; ERF, ethylene response factor; NAC, (NAM, ATAF, CUC); SPL, squamosa promoter binding protein-like; GRF, growth regulating factor; BT, BTB/TAZ; BBX, b-box protein; ARF, auxin response factor; LOB, lateral organ boundaries; BES1, BRI1-EMS-SUPPRESSOR 1; BR, brassinosteroid. The red dashed box represents the protein complex: MBW complex is constituted of three class of transcription factors (TFs), MYB, bHLH and WD40, while NF-Y complex is composed of TFs NF-YA, NF-YB, and NF-YC. TFs next to each other represent interaction of proteins.
Figure 3. Transcriptional regulation of flavonoid biosynthesis in plants. Abbreviations are as follows: MYB, v-myb avian myeloblastosis viral oncogene homolog; bHLH, basic helix-loop-helix; NF-Y, nuclear factor Y; ERF, ethylene response factor; NAC, (NAM, ATAF, CUC); SPL, squamosa promoter binding protein-like; GRF, growth regulating factor; BT, BTB/TAZ; BBX, b-box protein; ARF, auxin response factor; LOB, lateral organ boundaries; BES1, BRI1-EMS-SUPPRESSOR 1; BR, brassinosteroid. The red dashed box represents the protein complex: MBW complex is constituted of three class of transcription factors (TFs), MYB, bHLH and WD40, while NF-Y complex is composed of TFs NF-YA, NF-YB, and NF-YC. TFs next to each other represent interaction of proteins.
4. Perspectives
- Perspectives
Flavonoids are abundantly present in land plants where they have diverse functions; as dietary components, they also exert a variety of beneficial effects in humans [2][16][36][37][2, 16, 36, 37]. Elucidating the pathways involved in the biosynthesis of flavonoids will aid in better understanding their functions and potential uses. For example, the heterologous transformation of F3′5′H from Campanula medium (Canterbury bells) and A3′5′GT (anthocyanin 3′,5′-O-glucosyltransferase gene) from Clitoria ternatea (butterfly pea) driven by the native (Chrysanthemum morifolium) F3H promoter induced the synthesis of delphinidin and generated true blue Chrysanthemums [3][6][38][3, 6, 38]. Flavonoids have also been produced for food and medicine in engineered bacteria. The functional expression of plant-derived F3H, FLS, and OMT in Corynebacterium glutamicum yielded pterostilbene, kaempferol, and quercetin at high concentrations and purity [39]. In Escherichia coli, cyanidin 3-O-glucoside was generated through the induction of ANS and 3GT using a bicistronic expression cassette [40]. These observations highlight the important application and economic value of deciphering the pathways involved in flavonoid biosynthesis.
Over the past few decades, flavonoid biosynthesis has been among the most intensively investigated secondary metabolic pathways in plant biology, and a considerable number of studies have contributed to revealing the exquisite mechanisms underlying the biosynthesis of flavonoids in plants [1]. However, several questions remain outstanding. For example, no comprehensive model exists as yet regarding which enzymes catalyze the formation of 3-deoxyanthocyanidin; additionally, the biosynthesis of phlobaphenes needs to be further improved.
Plants are rich in diversity and often produce specific secondary metabolites. Recent studies have identified a unique flavone synthesis pathway in the root of the medicinal plant S. baicalensis, which generated root-specific flavones such as baicalein and norwogonin [41][42][41,42]. Accordingly, whether specific flavonoid biosynthesis pathways and metabolites also exist in other plants warrants further investigation, so as to continuously improve our knowledge of the flavonoid biosynthesis network.
In addition, combined multi-omics (genomics, transcriptomics, proteomics, and metabolomics) analysis provides a direction for the study of plant synthetic biology. In rice, a flavonoid 7-O-glycosyltransferase (OsUGT706C2) gene with a role in modulating flavonol (kaempferol) and flavone (luteolin and chrysoeriol) metabolism was identified by metabolite-based genome-wide association analysis [43]. Proteomics and transcriptomics, complemented with gas chromatography-mass spectrometry (GC-MS) analysis, aided in elucidating the flavonoid metabolic pathway during seed ripening in Camellia oleifera [44]. The constantly evolving multi-omics technology combined with big data analysis will likely lead to the identification of novel flavonoids and increased knowledge of the flavonoid biosynthesis network.
References
- Dong, N. Q.; Lin, H. X., Contribution of phenylpropanoid metabolism to plant development and plant-environment interactions. Journal of integrative plant biology 2020.
- Nabavi, S. M.; Šamec, D.; Tomczyk, M.; Milella, L.; Russo, D.; Habtemariam, S.; Suntar, I.; Rastrelli, L.; Daglia, M.; Xiao, J.; Giampieri, F.; Battino, M.; Sobarzo-Sanchez, E.; Nabavi, S. F.; Yousefi, B.; Jeandet, P.; Xu, S.; Shirooie, S., Flavonoid biosynthetic pathways in plants: Versatile targets for metabolic engineering. Biotechnology advances 2018, 38, 107316.
- Sasaki, N.; Nakayama, T., Achievements and Perspectives in Biochemistry Concerning Anthocyanin Modification for Blue Flower Coloration. Plant and Cell Physiology 2015, 56, 28-40.
- Winkel-Shirley, B., Flavonoid Biosynthesis. A Colorful Model for Genetics, Biochemistry, Cell Biology, and Biotechnology. Plant Physiology 2001, 126, 485-493.
- Tanaka, Y.; Brugliera, F.; Chandler, S., Recent Progress of Flower Colour Modification by Biotechnology. International Journal of Molecular Sciences 2009, 10, 5350-5369.
- Noda, N.; Yoshioka, S.; Kishimoto, S.; Nakayama, M.; Douzono, M.; Tanaka, Y.; Aida, R., Generation of blue chrysanthemums by anthocyanin B-ring hydroxylation and glucosylation and its coloration mechanism. Science Advances 2017, 3, e1602785.
- Sun, C.; Zhang, M.; Dong, H.; Liu, W.; Guo, L.; Wang, X., A spatially-resolved approach to visualize the distribution and biosynthesis of flavones in Scutellaria baicalensis Georgi. Journal of pharmaceutical and biomedical analysis 2020, 179, 113014.
- Grotewold, E., The genetics and biochemistry of floral pigments. Annual Review of Plant Biology 2006, 57, 761-780.
- Cavaiuolo, M.; Cocetta, G.; Ferrante, A., The Antioxidants Changes in Ornamental Flowers during Development and Senescence. Antioxidants 2013, 2, 132-155.
- Iwashina, T., Flavonoid Function and Activity to Plants and Other Organisms. Biological Sciences in Space 2003, 17, 24-44.
- Zhang, P.; Du, H.; Wang, J.; Pu, Y.; Yang, C.; Yan, R.; Yang, H.; Cheng, H.; Yu, D., Multiplex CRISPR/Cas9-mediated metabolic engineering increases soya bean isoflavone content and resistance to soya bean mosaic virus. Plant Biotechnology Journal 2020, 18, 1384-1395.
- Pourcel, L.; Routaboul, J. M.; Cheynier, V.; Lepiniec, L.; Debeaujon, I., Flavonoid oxidation in plants: from biochemical properties to physiological functions. Trends in plant science 2007, 12, 29-36.
- Tan, H.; Man, C.; Xie, Y.; Yan, J.; Chu, J.; Huang, J., A Crucial Role of GA-Regulated Flavonol Biosynthesis in Root Growth of Arabidopsis. Molecular plant 2019, 12, 521-537.
- Selvakumar, P.; Badgeley, A.; Murphy, P.; Anwar, H.; Sharma, U.; Lawrence, K.; Lakshmikuttyamma, A., Flavonoids and Other Polyphenols Act as Epigenetic Modifiers in Breast Cancer. Nutrients 2020, 12, 761.
- Imran, M.; Rauf, A.; Abu-Izneid, T.; Nadeem, M.; Shariati, M. A.; Khan, I. A.; Imran, A.; Orhan, I. E.; Rizwan, M.; Atif, M.; Gondal, T. A.; Mubarak, M. S., Luteolin, a flavonoid, as an anticancer agent: A review. Biomedicine & Pharmacotherapy 2019, 112, 108612.
- Fernandes, I.; Pérez-Gregorio, R.; Soares, S.; Mateus, N.; de Freitas, V., Wine Flavonoids in Health and Disease Prevention. Molecules 2017, 22, 292.
- Xu, W.; Dubos, C.; Lepiniec, L., Transcriptional control of flavonoid biosynthesis by MYB-bHLH-WDR complexes. Trends in plant science 2015, 20, 176-185.
- Dubos, C.; Stracke, R.; Grotewold, E.; Weisshaar, B.; Martin, C.; Lepiniec, L., MYB transcription factors in Arabidopsis. Trends in plant science 2010, 15, 573-581.
- Cao, Y.; Jia, H.; Xing, M.; Jin, R.; Grierson, D.; Gao, Z.; Sun, C.; Chen, K.; Xu, C.; Li, X., Genome-Wide Analysis of MYB Gene Family in Chinese Bayberry (Morella rubra) and Identification of Members Regulating Flavonoid Biosynthesis. Frontiers in plant science 2021, 12, 691384.
- Zhang, B.; Xu, X.; Huang, R.; Yang, S.; Li, M.; Guo, Y., CRISPR/Cas9-mediated targeted mutation reveals a role for AN4 rather than DPL in regulating venation formation in the corolla tube of Petunia hybrida. Horticulture research 2021, 8, 116.
- Li, J.; Luan, Q.; Han, J.; Zhang, C.; Liu, M.; Ren, Z., CsMYB60 directly and indirectly activates structural genes to promote the biosynthesis of flavonols and proanthocyanidins in cucumber. Horticulture research 2020, 7, 103.
- Xu, H.; Yang, G.; Zhang, J.; Wang, Y.; Zhang, T.; Wang, N.; Jiang, S.; Zhang, Z.; Chen, X., Overexpression of a repressor MdMYB15L negatively regulates anthocyanin and cold tolerance in red-fleshed callus. Biochemical and biophysical research communications 2018, 500, 405-410.
- Li, C.; Qiu, J.; Ding, L.; Huang, M.; Huang, S.; Yang, G.; Yin, J., Anthocyanin biosynthesis regulation of DhMYB2 and DhbHLH1 in Dendrobium hybrids petals. Plant Physiology and Biochemistry 2017, 112, 335-345.
- Totsuka, A.; Okamoto, E.; Miyahara, T.; Kouno, T.; Cano, E. A.; Sasaki, N.; Watanabe, A.; Tasaki, K.; Nishihara, M.; Ozeki, Y., Repressed expression of a gene for a basic helix-loop-helix protein causes a white flower phenotype in carnation. Breeding science 2018, 68, 139-143.
- Gonzalez, A.; Zhao, M.; Leavitt, J. M.; Lloyd, A. M., Regulation of the anthocyanin biosynthetic pathway by the TTG1/bHLH/Myb transcriptional complex in Arabidopsis seedlings. The Plant Journal 2008, 53, 814-827.
- Gao, Y.; Liu, J.; Chen, Y.; Tang, H.; Wang, Y.; He, Y.; Ou, Y.; Sun, X.; Wang, S.; Yao, Y., Tomato SlAN11 regulates flavonoid biosynthesis and seed dormancy by interaction with bHLH proteins but not with MYB proteins. Horticulture research 2018, 5, 27.
- Wang, J.; Li, G.; Li, C.; Zhang, C.; Cui, L.; Ai, G.; Wang, X.; Zheng, F.; Zhang, D.; Larkin, R. M.; Ye, Z.; Zhang, J., NF-Y plays essential roles in flavonoid biosynthesis by modulating histone modifications in tomato. The New phytologist 2020.
- Ni, J.; Bai, S.; Zhao, Y.; Qian, M.; Tao, R.; Yin, L.; Gao, L.; Teng, Y., Ethylene response factors Pp4ERF24 and Pp12ERF96 regulate blue light-induced anthocyanin biosynthesis in 'Red Zaosu' pear fruits by interacting with MYB114. Plant molecular biology 2019, 99, 67-78.
- Zhao, X.; Zeng, X.; Lin, N.; Yu, S.; Fernie, A. R.; Zhao, J., CsbZIP1-CsMYB12 mediates the production of bitter-tasting flavonols in tea plants (Camellia sinensis) through a coordinated activator-repressor network. Horticulture research 2021, 8, 110.
- Zhou, H.; Lin-Wang, K.; Wang, H.; Gu, C.; Dare, A. P.; Espley, R. V.; He, H.; Allan, A. C.; Han, Y., Molecular genetics of blood-fleshed peach reveals activation of anthocyanin biosynthesis by NAC transcription factors. The Plant Journal 2015, 82, 105-121.
- Li, C.; Wu, J.; Hu, K. D.; Wei, S. W.; Sun, H. Y.; Hu, L. Y.; Han, Z.; Yao, G. F.; Zhang, H., PyWRKY26 and PybHLH3 cotargeted the PyMYB114 promoter to regulate anthocyanin biosynthesis and transport in red-skinned pears. Horticulture research 2020, 7, 37.
- Ren, Y. R.; Zhao, Q.; Yang, Y. Y.; Zhang, T. E.; Wang, X. F.; You, C. X.; Hao, Y. J., The apple 14-3-3 protein MdGRF11 interacts with the BTB protein MdBT2 to regulate nitrate deficiency-induced anthocyanin accumulation. Horticulture research 2021, 8, 22.
- Luo, D.; Xiong, C.; Lin, A.; Zhang, C.; Sun, W.; Zhang, J.; Yang, C.; Lu, Y.; Li, H.; Ye, Z.; He, P.; Wang, T., SlBBX20 interacts with the COP9 signalosome subunit SlCSN5-2 to regulate anthocyanin biosynthesis by activating SlDFR expression in tomato. Horticulture research 2021, 8, 163.
- Wang, Y.; Wang, N.; Xu, H.; Jiang, S.; Fang, H.; Zhang, T.; Su, M.; Xu, L.; Zhang, Z.; Chen, X., Nitrogen Affects Anthocyanin Biosynthesis by Regulating MdLOB52 Downstream of MdARF19 in Callus Cultures of Red-Fleshed Apple (Malus sieversii f. niedzwetzkyana). Journal of Plant Growth Regulation 2017.
- Liang, T.; Shi, C.; Peng, Y.; Tan, H.; Xin, P.; Yang, Y.; Wang, F.; Li, X.; Chu, J.; Huang, J.; Yin, Y.; Liu, H., Brassinosteroid-Activated BRI1-EMS-SUPPRESSOR 1 Inhibits Flavonoid Biosynthesis and Coordinates Growth and UV-B Stress Responses in Plants. The Plant cell 2020, 32, 3224-3239.
- Jiao, C.; Sorensen, I.; Sun, X.; Sun, H.; Behar, H.; Alseekh, S.; Philippe, G.; Palacio Lopez, K.; Sun, L.; Reed, R.; Jeon, S.; Kiyonami, R.; Zhang, S.; Fernie, A. R.; Brumer, H.; Domozych, D. S.; Fei, Z.; Rose, J. K. C., The Penium margaritaceum Genome: Hallmarks of the Origins of Land Plants. Cell 2020, 181, 1097-1111.
- Lu, C.; Li, Y.; Cui, Y.; Ren, J.; Qi, F.; Qu, J.; Huang, H.; Dai, S., Isolation and Functional Analysis of Genes Involved in Polyacylated Anthocyanin Biosynthesis in Blue Senecio cruentus. Frontiers in plant science 2021, 12, 640746.
- Noda, N.; Aida, R.; Kishimoto, S.; Ishiguro, K.; Fukuchi-Mizutani, M.; Tanaka, Y.; Ohmiya, A., Genetic engineering of novel bluer-colored chrysanthemums produced by accumulation of delphinidin-based anthocyanins. Plant and Cell Physiology 2013, 54, 1684-1695.
- Kallscheuer, N.; Vogt, M.; Bott, M.; Marienhagen, J., Functional expression of plant-derived O-methyltransferase, flavanone 3-hydroxylase, and flavonol synthase in Corynebacterium glutamicum for production of pterostilbene, kaempferol, and quercetin. Journal of biotechnology 2017, 258, 190-196.
- Lim, C. G.; Wong, L.; Bhan, N.; Dvora, H.; Xu, P.; Venkiteswaran, S.; Koffas, M. A., Development of a Recombinant Escherichia coli Strain for Overproduction of the Plant Pigment Anthocyanin. Applied and environmental microbiology 2015, 81, 6276-6284.
- Zhao, Q.; Yang, J.; Cui, M. Y.; Liu, J.; Fang, Y.; Yan, M.; Qiu, W.; Shang, H.; Xu, Z.; Yidiresi, R.; Weng, J. K.; Pluskal, T.; Vigouroux, M.; Steuernagel, B.; Wei, Y.; Yang, L.; Hu, Y.; Chen, X. Y.; Martin, C., The Reference Genome Sequence of Scutellaria baicalensis Provides Insights into the Evolution of Wogonin Biosynthesis. Molecular plant 2019, 12, 935-950.
- Zhao, Q.; Cui, M. Y.; Levsh, O.; Yang, D.; Liu, J.; Li, J.; Hill, L.; Yang, L.; Hu, Y.; Weng, J. K.; Chen, X. Y.; Martin, C., Two CYP82D Enzymes Function as Flavone Hydroxylases in the Biosynthesis of Root-Specific 4'-Deoxyflavones in Scutellaria baicalensis. Molecular plant 2018, 11, 135-148.
- Zhang, F.; Guo, H.; Huang, J.; Yang, C.; Li, Y.; Wang, X.; Qu, L.; Liu, X.; Luo, J., A UV-B-responsive glycosyltransferase, OsUGT706C2, modulates flavonoid metabolism in rice. Science China. Life sciences 2020, 63, 1037-1052.
- Ye, Z.; Yu, J.; Yan, W.; Zhang, J.; Yang, D.; Yao, G.; Liu, Z.; Wu, Y.; Hou, X., Integrative iTRAQ-based proteomic and transcriptomic analysis reveals the accumulation patterns of key metabolites associated with oil quality during seed ripening of Camellia oleifera. Horticulture research 2021, 8, 157.
