Your browser does not fully support modern features. Please upgrade for a smoother experience.
Please note this is a comparison between Version 2 by Yvaine Wei and Version 1 by Tarun Kumar Upadhyay.
SARS-CoV-2 is attached to host cells via binding to the viral spike (S) proteins and its cellular receptors angiotensin-converting enzyme 2 (ACE2). Consequently, the S protein is primed with serine proteases TMPRSS2 and TMPRSS4, which facilitate the fusion of viral and cellular membranes result in the entry of viral RNA into the host cell. The long-term protective immunity is provided by the vaccine antigen (or pathogen)-specific immune effectors and the activation of immune memory cells that can be efficiently and rapidly reactivated upon pathogen exposure.
- SARS-CoV-2
- adenoviral vector-based vaccine
- Vaccine
- Immunity
1. Sensing of SARS-CoV-2 Pathogen by Innate Immunity
As an initial line of protection, vertebrates use innate immunological responses as their defensive systems. The innate immune response performs three primary functions: (i) limiting viral replication within infected cells, (ii) inducing an antiviral state in the immediate tissue environment, along with the recruitment of innate immune effector cells, and (iii) priming the adaptive immune response [10,11][1][2]. Pathogen recognition receptors (PRRs) distinguish microbial components known as pathogen-associated molecular patterns that are likely to be required for the survival of the microorganism. PRRs are expressed constitutively in the host and recognize infections or their intermediate products throughout the pathogen’s life cycle. PRRs react with distinct microbial components different from the self and activate specific signaling pathways that drive biological responses against pathogens [12,13][3][4]. In the case of RNA viruses, the mammalian host’s innate immune response has two primary routes for controlling viral infections. One includes signaling pathways mediated by TLR 3, 7 or 9 in response to detection of the viral DNA or its intermediates. The other is carried out by cytoplasmic RNA helicases such as melanoma differentiation-associated gene 5 (MDA5) or retinoic acid-inducible gene I (RIG I), which may detect 5′ triphosphorylated and double viral RNAs as well as stimulate the host antiviral innate immune system [14,15][5][6]. Several observations highlight the critical role of innate immunity in SARS-CoV-2 regulation. Human coronaviruses such as SARS, MERS, and SARS-CoV-2 have acquired specific immune-suppressive mechanisms such as a part of the viruses’ genetic material is committed to code for the proteins that particularly target human innate immunity systems, prominently IFN response pathways by blocking TLR3/7 signaling [16,17][7][8]. The innate immune system also triggers activates the adaptive immune responses, which act together just to eradicate infections [11][2]. However, adaptive immune responses are more specific and provide long-term protection from reinfection with the same type of pathogen [10][1].
2. Humoral and Cell Mediated Immune Responses against SARS-CoV-2
Human SARS-CoV-2 infection appears to include both humoral as well as cell-mediated immunity [18,19][9][10]. Antibody molecules produced by plasma cells mediate the humoral immune response. When antigen attaches to the B-cell antigen receptor, it alerts B cells while also being internalized and processed into peptides that activate the armed helper [20][11]. In cell-mediated immune responses, the second kind of adaptive immune response activated T cells directly respond to a foreign antigen presented to them on the surface of a host cell. For example, a virus-infected host cell bearing viral antigens on its surface may be destroyed by a T cell, eliminating the infected cell before the virus has a chance to reproduce. In other cases, the T cell secretes signal molecules that stimulate macrophages to destroy the intruders they have phagocytized [10][1]. Adaptive immunity involves the coordination of T and B cell immune responses to the SARS-CoV-2 virus. Immune responses to the SARS virus begin within 7–10 days after infection [21][12]. Early responses to COVID-19 infection include IgM and IgA, although it is unclear whether they can alter the course of the illness [22,23,24][13][14][15]. First, early IgG responses emerge from germinal centers after T follicular cells stimulate naïve B cells, which develop into activated B cells, which then differentiate into B memory cells and IgG generating plasmablasts. B memory cells and long-lived plasma cells in the bone marrow can reactivate antigen-specific responses against SARS-CoV-2 pathogen if exposed again. Moreover, this overlooks the importance of T cell memory for COVID-19 antigenic determinates, which can lead to efficient cytotoxic T cell immunity and aid in B cell responses [25][16].
3. Vaccine-Induced Immune Responses against SARS-CoV-2 Infections
3.1. Nucleic Acid-Based Vaccines for COVID-19
The usage of nucleic acid-based vaccines is a new method of vaccination that induces immune responses comparable to those elicited by live, attenuated vaccines. When nucleic acid vaccines are administered, the endogenous production of viral proteins having native conformation, glycosylation patterns, and other posttranslational modifications that match antigen generated during normal viral infection occurs. To date various protein antigens, nucleic acid vaccines have been demonstrated to elicit both antibody and cytotoxic T-lymphocyte responses. The convenience of the vector, ease of administration, the longevity of expression, and evidence of integration are the advantages of nucleic acid-based vaccines. More research is needed to determine the practicality, safety, and efficacy of this novel and promising technique [26][17]. Nucleic acid vaccines, at their most basic, use the host’s transcriptional and translational machinery to create the target gene product. This polypeptide product is then identified by immune system components. The initial research focused on the absorption of plasmid DNA by myocytes. Despite the fact that myocytes can deliver antigen to immune cells, they are not the major activators of immune cells. An immune response is instead largely initiated by antigen presentation cells (APC) include dendritic cells (DC), B cells, macrophages, and Langerhans cells [27,28][18][19]. The most potent APC, dendritic cells acquire antigen via three major mechanisms. First, nucleic acid vaccines may be directly transfected into DC. Second, DC takes up soluble antigen produced or released by transfected cells from interstitial areas. Third, and probably most intriguingly, DC preferentially takes up damaged or dead cells as a result of the vaccination or its action [29][20].
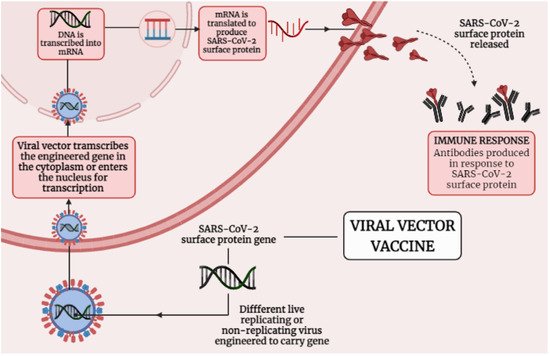
3.2. Immunological Mechanisms of Different Adenoviral Vector-Based Vaccines-Induced Protection against SARS-CoV-2
Adenoviral vectors were used with poxviral and DNA vectors to enhance immunogenicity, with eitheradenovirus or modified vaccinia virus Ankara prime-boost regimens improving both cellular and humoral responses [28][19]. Figure 3 1 presenting the mechanism that how viral vector-based vaccine works against SARS-CoV-2. Adenoviral vectors have been investigated as a platform for carrying and expressing a range of transgenes as a foundation for vaccine development [37][21]. The ChAdOx1 nCoV-19 adenoviral vector-based vaccine (AZD1222) was constructed at Oxford University and consists of simian adenovirus vector ChAdOx1, which carries the full-pace structural surface glycoprotein (spike protein) of theSARS-CoV-2. ChAdOx1 nCoV-19 encodes a spike protein with a codon-optimized coding sequence [28,54,63][19][22][23]. ChAdOx1 nCoV-19 elicits a widespread and strong T cell response to both S antigen components. After vaccination, there was a significant increase in B cell activation and proliferation, and anti-IgA and IgG antibodies to the SARS-CoV-2 spike protein were easily identified in sera from vaccinated individuals [55][24]. Analyses of cytokine secretion after peptide stimulation of PBMCs revealed that IFN- and IL-2 production were higher in those who got the ChAdOx1 vaccination compared to controls, while IL-4 and IL-13 levels were not. Similarly, flow cytometry phenotyping revealed that CD4+ T cells produced primarily Th1 cytokines (IFN-, IL-2, and TNF-) rather than Th2 cytokines (IL-5 and IL-13). Importantly, it showed that immunization with ChAdOx1 nCoV-19 generates a mainly Th1 response using a variety of methods (multiplex cytokine profiling, ICS analysis, and antibody isotype profiling). In a phase 1/2 research, a single dosage of ChAdOx1 nCoV-19 resulted in a rise in spike-specific antibodies by day 28 and neutralizing antibodies in all participants after a booster dose. After vaccination, there was a significant increase in B cell activation and proliferation, and anti-IgA and IgG antibodies to the SARS-CoV-2 spike protein were easily identified in sera from vaccinated participants [28][19]. T-cell responses believed to play an important role in COVID-19 mitigation; persons who have been treated but asymptomatic developed a robust memory T-cell response in the absenteeism of clinical disease, despite the lack of a recognizable humoral response [29,52][20][25]. ChAdOx1 nCoV-19 was shown to be safe, tolerable, and immunogenic, with reactogenicity decreased by paracetamol. A sole dose elicited both humoral and cellular responses against SARS-CoV-2, and just a booster dose increased neutralizing antibody titers [28,53][19][26].
Figure 31. Shows the mechanism of Viral vector-based vaccine against SARS-CoV-2.
4. Vaccines and Its Role in Inducing Humoral Adaptive Immunity
Recent research has revealed that the novel SARS-CoV-2 virus employs a similar mechanism for cell entrance [70][27]. To connect to host cells, the viral S protein attaches to the angiotensin-converting enzyme 2 (ACE2), the viral receptor. The S protein is then primed by host cell proteases, furin, and the serine proteases TMPRSS2 and TMPRSS4, allowing viral and cellular membranes to fuse and viral RNA to enter the host cell [71][28]. Long-term protective immunity is supplied by vaccination antigen (or pathogen)-specific immune effectors and the activation of immunological memory cells that can be effectively and quickly reactivated in the event of pathogen exposure [72,73,74][29][30][31]. Most vaccines that have been approved thus far stimulate antibodies generated by B cells, which are believed to be responsible for the vaccine’s long-term protection [75,76][32][33]. Vaccine antigen and pathogen binding to B cell receptors (antibody in membrane-bound form) induces the production of an initial activation marker CD69 and also a chemokine receptor CCR7 that drives antigen-specific B cells onto secondary lymphoid tissue T cell zones [77,78,79,80][34][35][36][37]. Vaccine antigen-specific B cells are likely to engage with newly activated T cells and DCs, particularly follicular DCs with specific surface molecules, at this site (CD40, CD80, CD86). This T cell assistance accelerates B cell development into antibody-secreting, short-lived plasma cells that generate low-affinity germ-line encoded antibodies [80][37]. The development of neutralizing antibodies aimed towards spike protein is a key component of successful vaccination. This is the foundation for many clinical trials, as well as the creation of monoclonal antibody cocktails, which have proven crucial in COVID-19 treatments in the absence of a vaccine. However, nothing was known until recently about what constituted a successful immunological response to COVID-19. The architecture of antibodies targeting the SARS-CoV-2 receptor-binding domain (RBD) is an essential factor. Recent studies have demonstrated that antibodies bound to the receptor-binding domain RBD are important for long-term protective immunity against COVID-19 infection and are linked with improved patient survival [81,82,83,84][38][39][40][41]. Virus-neutralizing antibodies are largely responsible for the protection provided by presently available vaccinations. These antibodies often inhibit the virus’s contact with its cellular receptor or prevent the virus from undergoing the conformational changes necessary for fusion with the cell membrane. Vaccination’s objective is to generate long-term protective immunity, which is a feature of adaptive immunity.
References
- The Adaptive Immune System. In Molecular Biology of the Cell, 4th ed.; Garland Science: New York, NY, USA, 2002. Available online: https://www.ncbi.nlm.nih.gov/books/NBK21070/ (accessed on 2 September 2021).
- Sette, A.; Crotty, S. Adaptive immunity to SARS-CoV-2 and COVID-19. Cell 2021, 184, 861–880.
- Akira, S.; Uematsu, S.; Takeuchi, O. Pathogen recognition and innate immunity. Cell 2006, 124, 783–801.
- Pulendran, B. Modulating vaccine responses with dendritic cells and Toll-like receptors. Immunol. Rev. 2004, 199, 227–250.
- Loo, Y.M.; Fornek, J.; Crochet, N.; Bajwa, G.; Perwitasari, O.; Martinez-Sobrido, L.; Akira, S.; Gill, M.A.; García-Sastre, A.; Katze, M.G.; et al. Distinct RIG-I and MDA5 signaling by RNA viruses in innate immunity. J. Virol. 2008, 82, 335–345.
- Takeshita, F.; Kobiyama, K.; Miyawaki, A.; Jounai, N.; Okuda, K. The non-canonical role of Atg family members as suppressors of innate antiviral immune signaling. Autophagy 2008, 4, 67–69.
- Lei, X.; Dong, X.; Ma, R.; Wang, W.; Xiao, X.; Tian, Z.; Wang, C.; Wang, Y.; Li, L.; Ren, L.; et al. Activation and evasion of type I interferon responses by SARS-CoV-2. Nat. Commun. 2020, 11, 3810.
- Young, B.E.; Fong, S.W.; Chan, Y.H.; Mak, T.M.; Ang, L.W.; Anderson, D.E.; Lee, C.Y.; Amrun, S.N.; Lee, B.; Goh, Y.S.; et al. Effects of a major deletion in the SARS-CoV-2 genome on the severity of infection and the inflammatory response: An observational cohort study. Lancet 2020, 396, 603–611.
- Gudbjartsson, D.F.; Norddahl, G.L.; Melsted, P.; Gunnarsdottir, K.; Holm, H.; Eythorsson, E.; Arnthorsson, A.O.; Helgason, D.; Bjarnadottir, K.; Ingvarsson, R.F.; et al. Humoral Immune Response to SARS-CoV-2 in Iceland. N. Engl. J. Med. 2020, 383, 1724–1734.
- Chen, G.; Wu, D.; Guo, W.; Cao, Y.; Huang, D.; Wang, H.; Wang, T.; Zhang, X.; Chen, H.; Yu, H.; et al. Clinical and immunological features of severe and moderate coronavirus disease 2019. J. Clin. Investig. 2020, 130, 2620–2629.
- The Humoral Immune Response. In Immunobiology: The Immune System in Health and Disease, 5th ed.; Garland Science: New York, NY, USA, 2001. Available online: https://www.ncbi.nlm.nih.gov/books/NBK10752/ (accessed on 1 September 2021).
- Jordan, S.C. Innate and adaptive immune responses to SARS-CoV-2 in humans: Relevance to acquired immunity and vaccine responses. Clin. Exp. Immunol. 2021, 204, 310–320.
- Stephens, D.S.; McElrath, M.J. COVID-19 and the Path to Immunity. JAMA 2020, 324, 1279–1281.
- Suthar, M.S.; Zimmerman, M.G.; Kauffman, R.C.; Mantus, G.; Linderman, S.L.; Hudson, W.H.; Vanderheiden, A.; Nyhoff, L.; Davis, C.W.; Adekunle, O.; et al. Rapid Generation of Neutralizing Antibody Responses in COVID-19 Patients. Cell Rep. Med. 2020, 1, 100040.
- Robbiani, D.F.; Gaebler, C.; Muecksch, F.; Lorenzi, J.; Wang, Z.; Cho, A.; Agudelo, M.; Barnes, C.O.; Gazumyan, A.; Finkin, S.; et al. Convergent antibody responses to SARS-CoV-2 in convalescent individuals. Nature 2020, 584, 437–442.
- Dan, J.M.; Mateus, J.; Kato, Y.; Hastie, K.M.; Yu, E.D.; Faliti, C.E.; Grifoni, A.; Ramirez, S.I.; Haupt, S.; Frazier, A.; et al. Immunological memory to SARS-CoV-2 assessed for up to 8 months after infection. Science 2021, 371.
- Rauch, S.; Jasny, E.; Schmidt, K.E.; Petsch, B. New Vaccine Technologies to Combat Outbreak Situations. Front. Immunol. 2018, 9, 1963.
- Corr, M.; Lee, D.J.; Carson, D.A.; Tighe, H. Gene vaccination with naked plasmid DNA: Mechanism of CTL priming. J. Exp. Med. 1996, 184, 1555–1560.
- Folegatti, P.M.; Ewer, K.J.; Aley, P.K.; Angus, B.; Becker, S.; Belij-Rammerstorfer, S.; Bellamy, D.; Bibi, S.; Bittaye, M.; Clutterbuck, E.A.; et al. Safety and immunogenicity of the ChAdOx1 nCoV-19 vaccine against SARS-CoV-2: A preliminary report of a phase 1/2, single-blind, randomised controlled trial. Lancet 2020, 396, 467–478.
- Porgador, A.; Irvine, K.R.; Iwasaki, A.; Barber, B.H.; Restifo, N.P.; Germain, R.N. Predominant role for directly transfected dendritic cells in antigen presentation to CD8+ T cells after gene gun immunization. J. Exp. Med. 1998, 188, 1075–1082.
- Lindsay, K.E.; Bhosle, S.M.; Zurla, C.; Beyersdorf, J.; Rogers, K.A.; Vanover, D.; Xiao, P.; Araínga, M.; Shirreff, L.M.; Pitard, B.; et al. Visualization of early events in mRNA vaccine delivery in non-human primates via PET-CT and near-infrared imaging. Nat. Biomed. Eng. 2019, 3, 371–380.
- Liu, M.A. DNA vaccines: An historical perspective and view to the future. Immunol. Rev. 2011, 239, 62–84.
- Ramasamy, M.N.; Minassian, A.M.; Ewer, K.J.; Flaxman, A.L.; Folegatti, P.M.; Owens, D.R.; Voysey, M.; Aley, P.K.; Angus, B.; Babbage, G.; et al. Safety and immunogenicity of ChAdOx1 nCoV-19 vaccine administered in a prime-boost regimen in young and old adults (COV002): A single-blind, randomised, controlled, phase 2/3 trial. Lancet 2021, 396, 1979–1993.
- Schirmbeck, R.; Böhm, W.; Reimann, J. DNA vaccination primes MHC class I-restricted, simian virus 40 large tumor antigen-specific CTL in H-2d mice that reject syngeneic tumors. J. Immunol. 1996, 157, 3550–3558.
- Leitner, W.W.; Ying, H.; Restifo, N.P. DNA and RNA-based vaccines: Principles, progress and prospects. Vaccine 1999, 18, 765–777.
- Tang, D.C.; DeVit, M.; Johnston, S.A. Genetic immunization is a simple method for eliciting an immune response. Nature 1992, 356, 152–154.
- Hoffmann, M.; Kleine-Weber, H.; Schroeder, S.; Krüger, N.; Herrler, T.; Erichsen, S.; Schiergens, T.S.; Herrler, G.; Wu, N.H.; Nitsche, A.; et al. SARS-CoV-2 Cell Entry Depends on ACE2 and TMPRSS2 and Is Blocked by a Clinically Proven Protease Inhibitor. Cell 2020, 181, 271–280.
- Zang, R.; Gomez Castro, M.F.; McCune, B.T.; Zeng, Q.; Rothlauf, P.W.; Sonnek, N.M.; Liu, Z.; Brulois, K.F.; Wang, X.; Greenberg, H.B.; et al. TMPRSS2 and TMPRSS4 promote SARS-CoV-2 infection of human small intestinal enterocytes. Sci. Immunol. 2020, 5, 1–10.
- Kalia, V.; Sarkar, S.; Gourley, T.S.; Rouse, B.T.; Ahmed, R. Differentiation of memory B and T cells. Curr. Opin. 2006, 18, 255–264.
- Quan, F.S.; Huang, C.; Compans, R.W.; Kang, S.M. Virus-like particle vaccine induces protective immunity against homologous and heterologous strains of influenza virus. J. Virol. 2007, 81, 3514–3524.
- Quan, F.S.; Compans, R.W.; Nguyen, H.H.; Kang, S.M. Induction of heterosubtypic immunity to influenza virus by intranasal immunization. J. Virol. 2008, 82, 1350–1359.
- Crotty, S.; Aubert, R.D.; Glidewell, J.; Ahmed, R. Tracking human antigen-specific memory B cells: A sensitive and generalized ELISPOT system. J. Immunol. Methods 2004, 286, 111–122.
- Rappuoli, R. Bridging the knowledge gaps in vaccine design. Nat. Biotechnol. 2007, 25, 1361–1366.
- Sancho, D.; Gómez, M.; Sánchez-Madrid, F. CD69 is an immunoregulatory molecule induced following activation. Trends Immunol. 2005, 26, 136–140.
- McHeyzer-Williams, L.J.; McHeyzer-Williams, M.G. Antigen-specific memory B cell development. Annu. Rev. Immunol. 2005, 23, 487–513.
- Goel, H.; Gupta, I.; Mourya, M.; Gill, S.; Chopra, A.; Ranjan, A.; Rath, G.K.; Tanwar, P.A. systematic review of clinical and laboratory parameters of 3000 COVID-19 cases. Obstet. Gynecol. Sci. 2021, 64, 174–189.
- Reif, K.; Ekland, E.H.; Ohl, L.; Nakano, H.; Lipp, M.; Förster, R.; Cyster, J.G. Balanced responsiveness to chemoattractants from adjacent zones determines B-cell position. Nature 2002, 416, 94–99.
- Kelsoe, G. Studies of the humoral immune response. Immunol. Res. 2000, 22, 199–210.
- Barnes, C.O.; Jette, C.A.; Abernathy, M.E.; Dam, K.A.; Esswein, S.R.; Gristick, H.B.; Malyutin, A.G.; Sharaf, N.G.; Huey-Tubman, K.E.; Lee, Y.E.; et al. SARS-CoV-2 neutralizing antibody structures inform therapeutic strategies. Nature 2020, 588, 682–687.
- Goel, H.; Goyal, K.; Baranwal, P.; Dixit, A.; Upadhyay, T.K.; Upadhye, V.J. The Diagnostics Technologies and Control of COVID-19. Lett. Appl. NanoBioSci. 2021, 11, 3120–3133.
- Secchi, M.; Bazzigaluppi, E.; Brigatti, C.; Marzinotto, I.; Tresoldi, C.; Rovere-Querini, P.; Poli, A.; Castagna, A.; Scarlatti, G.; Zangrillo, A.; et al. COVID-19 survival associates with the immunoglobulin response to the SARS-CoV-2 spike receptor binding domain. J. Clin. Investig. 2020, 130, 6366–6378.
More
