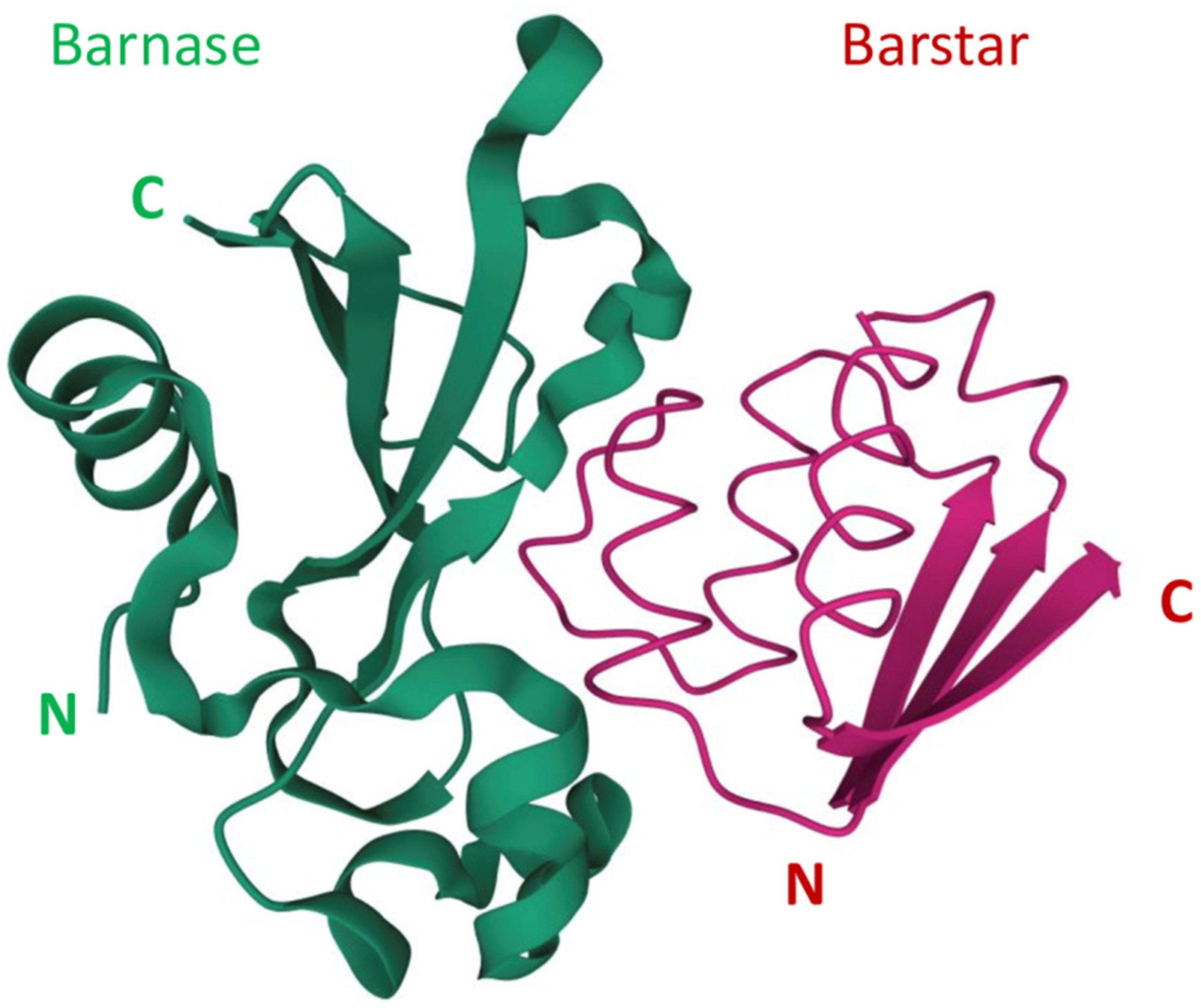
Barnase is an extracellular ribonuclease secreted by Bacillus amyloliquefaciens that was originally studied as a small stable enzyme with robust folding. The identification of barnase intracellular inhibitor barstar led to the discovery of an incredibly strong protein-protein interaction. Together, barnase and barstar provide a fully genetically encoded toxin-antitoxin pair having an extremely low dissociation constant. Moreover, compared to other dimerization systems, the barnase-barstar module provides the exact one-to-one ratio of the complex components and possesses high stability of each component in a complex and high solubility in aqueous solutions without self-aggregation. The unique properties of barnase and barstar allow the application of this pair for the engineering of different variants of targeted anticancer compounds and cytotoxic supramolecular complexes. Using barnase in suicide gene therapy has also found its niche in anticancer therapy.
- barnase
- barstar
- cancer therapy
- nanoparticles
1. Introduction
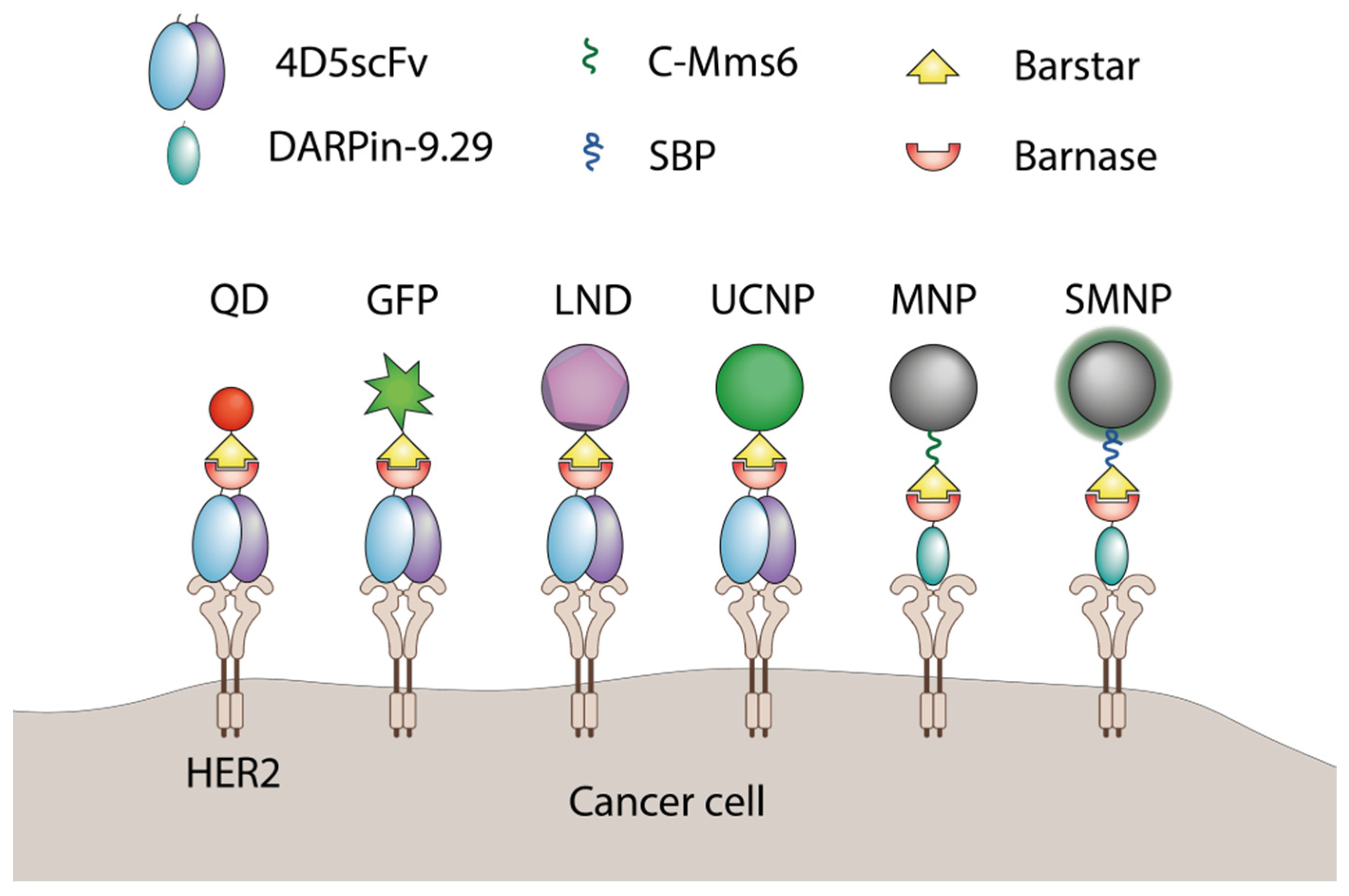
2. The Use of Barnase-Barstar Interaction for Supramolecular Complexes Assembly
3. Conclusions
References
- Hartley, R.W. Barnase and barstar: Two small proteins to fold and fit together. Trends Biochem. Sci. 1989, 14, 450–454.
- Nishimura, S.; Nomura, M. Ribonuclease of Bacillus subtilis. BBA-Biochim. Biophys. Acta 1958, 30, 430–431.
- Smeaton, J.R.; Elliott, W.H. Isolation and properties of a specific bacterial ribonuclease inhibitor. BBA Sect. Nucleic Acids Protein Synth. 1967, 145, 547–560.
- Hartley, R.W.; Rogerson, D.L. Production and Purification of the Extracellular Ribonuclease of Bacillus Amyloliquefaciens (Barnase) and Its Intracellular Inhibitor (Barstar) I. Barnase. Prep. Biochem. 1972, 2, 229–242.
- Hartley, R.W.; Rogerson, D.L.; Smeaton, J.R. Production and Purification of the Extracellular Ribonuclease of Bacillus Amyloliquefaciens (Barnase) and Its Intracellular Inhibitor (Barstar) II. Barstar. Prep. Biochem. 1972, 2, 243–250.
- Hartley, R.W.; Barker, E.A. Amino-acid sequence of extracellular ribonuclease (barnase) of bacillus amyloliquefaciens. Nat. New Biol. 1972, 235, 15–16.
- Hartley, R.T.; Smeaton, J.R. On the Reaction between the Extracellular Ribonuclease of Bacillus amyloliquefaciens (Barnase) and Its Intracellular Inhibitor (Barstar). J. Biol. Chem. 1973, 248, 5624–5626.
- Hartley, R.W. Barnase-barstar interaction. In Methods in Enzymology; Academic Press Inc.: New York, NY, USA, 2001; Volume 341, pp. 599–611.
- Deyev, S.M.; Waibel, R.; Lebedenko, E.N.; Schubiger, A.P.; Plückthun, A. Design of multivalent complexes using the barnase·barstar module. Nat. Biotechnol. 2003, 21, 1486–1492.
- Korchuganov, D.S.; Nolde, S.B.; Reibarkh, M.Y.; Orekhov, V.Y.; Schulga, A.A.; Ermolyuk, Y.S.; Kirpichnikov, M.P.; Arseniev, A.S. NMR study of monomer-dimer equilibrium of barstar in solution. J. Am. Chem. Soc. 2001, 123, 2068–2069.
- Paddon, C.J.; Hartley, R.W. Cloning, sequencing and transcription of an inactivated copy of Bacillus amyloliquefaciens extracellular ribonuclease (barnase). Gene 1985, 40, 231–239.
- Paddon, C.J.; Hartley, R.W. Expression of Bacillus amyloliquefaciens extracellular ribonuclease (barnase) in Escherichia coli following an inactivating mutation. Gene 1987, 53, 11–19.
- Hartley, R.W. Barnase and barstar. Expression of its cloned inhibitor permits expression of a cloned ribonuclease. J. Mol. Biol. 1988, 202, 913–915.
- Kellis, J.T.; Nyberg, K.; Šail, D.; Fersht, A.R. Contribution of hydrophobic interactions to protein stability. Nature 1988, 333, 784–786.
- Khurana, R.; Udgaonkar, J.B. Equilibrium Unfolding Studies of Barstar: Evidence for an Alternative Conformation Which Resembles a Molten Globule. Biochemistry 1994, 33, 106–115.
- Mossakowska, D.E.; Nyberg, K.; Fersht, A.R. Kinetic Characterization of the Recombinant Ribonuclease from Bacillus amyloliquefaciens (Barnase) and Investigation of Key Residues in Catalysis by Site-Directed Mutagenesis. Biochemistry 1989, 28, 3843–3850.
- Jones, D.N.M.; Bycroft, M.; Lubienski, M.J.; Fersht, A.R. Identification of the barstar binding site of barnase by NMR spectroscopy and hydrogen-deuterium exchange. FEBS Lett. 1993, 331, 165–172.
- Schreiber, G.; Fersht, A.R. Interaction of Barnase with Its Polypeptide Inhibitor Barstar Studied by Protein Engineering. Biochemistry 1993, 32, 5145–5150.
- Deyev, S.M.; Yazynin, S.A.; Kuznetsov, D.A.; Jukovich, M.; Hartley, R.W. Ribonuclease-charged vector for facile direct cloning with positive selection. Mol. Gen. Genet. 1998, 259, 379–382.
- Yazynin, S.; Lange, H.; Mokros, T.; Deyev, S.; Lemke, H. A new phagemid vector for positive selection of recombinants based on a conditionally lethal barnase gene. FEBS Lett. 1999, 452, 351–354.
- Mariani, C.; De Beuckeleer, M.; Truettner, J.; Leemans, J.; Goldberg, R.B. Induction of male sterility in plants by a chimaeric ribonuclease gene. Nature 1990, 347, 737–741.
- Goldman, M.H.S.; Goldberg, R.B.; Mariani, C. Female sterile tobacco plants are produced by stigma-specific cell ablation. EMBO J. 1994, 13, 2976–2984.
- Van Poucke, K.; Karimi, M.; Gheysen, G. Analysis of nematode-responsive promoters in sugar beet hairy roots. Meded. Rijksuniv. Gent. Fak. Landbouwkd Toegep. Biol. Wet 2001, 66, 591–598.
- Leuchtenberger, S.; Perz, A.; Gatz, C.; Bartsch, J.W. Conditional cell ablation by stringent tetracycline-dependent regulation of barnase in mammalian cells. Nucleic Acids Res. 2001, 29, 76.
- Ilinskaya, O.N.; Shah Mahmud, R. Ribonucleases as antiviral agents. Mol. Biol. 2014, 48, 615–623.
- Gribencha, S.V.; Potselueva, L.A.; Barinskiĭ, I.F.; Balandin, T.G.; Deev, S.M.; Leshchinskaia, I.B. The antiviral activity of RNAse Bacillus intermedius in experiments with mice preinfected with street rabies virus. Vopr. Virusol. 2004, 49, 38–41.
- Gribencha, S.V.; Potselueva, L.A.; Barinskiĭ, I.F.; Deev, S.M.; Balandin, T.G.; Leshchinskaia, I.B. Antiviral activity of Bacillus intermedius RNAase in guinea-pigs and rabbits infected with outdoor rabies virus. Vopr. Virusol. 2006, 51, 41–43.
- Glinka, E.M. Killing of cancer cells through the use of eukaryotic expression vectors harbouring genes encoding nucleases and ribonuclease inhibitor. Tumor Biol. 2015, 36, 3147–3157.
- Edelweiss, E.; Balandin, T.G.; Ivanova, J.L.; Lutsenko, G.V.; Leonova, O.G.; Popenko, V.I.; Sapozhnikov, A.M.; Deyev, S.M. Barnase as a new therapeutic agent triggering apoptosis in human cancer cells. PLoS ONE 2008, 3, e2434.
- Ulyanova, V.; Dudkina, E.; Nadyrova, A.; Kalashnikov, V.; Surchenko, Y.; Ilinskaya, O. The cytotoxicity of rnase-derived peptides. Biomolecules 2021, 11, 16.
- Garipov, A.R.; Nesmelov, A.A.; Cabrera-Fuentes, H.A.; Ilinskaya, O.N. Bacillus intermedius ribonuclease (BINASE) induces apoptosis in human ovarian cancer cells. Toxicon. 2014, 92, 54–59.
- Malhotra, V.; Perry, M.C. Classical Chemotherapy: Mechanisms, Toxicities and the Therapeutc Window. Cancer Biol. Ther. 2003, 2, S2–S4.
- Gotte, G.; Menegazzi, M. Biological Activities of Secretory RNases: Focus on Their Oligomerization to Design Antitumor Drugs. Front. Immunol. 2019, 10, 2626.
- Guillet, V.; Lapthorn, A.; Hartley, R.; Mauguen, Y. Recognition between a bacterial ribonuclease, barnase, and its natural inhibitor, barstar. Structure 1993, 1, 165–176.
- Buckle, A.M.; Schreiber, G.; Fersht, A.R. Protein-Protein Recognition: Crystal Structural Analysis of a Barnase-Barstar Complex at 2.0-Å Resolution. Biochemistry 1994, 33, 8878–8889.
- Aghayeva, U.F.; Nikitin, M.P.; Lukash, S.V.; Deyev, S.M. Denaturation-resistant bifunctional colloidal superstructures assembled via the proteinaceous barnase-barstar interface. ACS Nano 2013, 7, 950–961.
- Spång, H.C.; Braathen, R.; Bogen, B. Heterodimeric barnase-barstar vaccine molecules: Influence of one versus two targeting units specific for antigen presenting cells. PLoS ONE 2012, 7, e45393.
- Braathen, R.; Spång, H.C.L.; Hinke, D.M.; Blazevski, J.; Bobic, S.; Fossum, E.; Bogen, B. A DNA vaccine that encodes an antigen-presenting cell-specific heterodimeric protein protects against cancer and influenza. Mol. Ther. Methods Clin. Dev. 2020, 17, 378–392.
- Sapozhnikov, A.M.; Klinkova, A.V.; Shustova, O.A.; Grechikhina, M.V.; Kilyachus, M.S.; Stremovskiy, O.A.; Kovalenko, E.I.; Deyev, S.M. A Novel Approach to Anticancer Therapy: Molecular Modules Based on the Barnase:Barstar Pair for Targeted Delivery of HSP70 to Tumor Cells. Acta Nat. 2018, 10, 85–91.
- Kalinin, R.; Shipunova, V.; Chernikov, I.; Volkov, D.; Shulga, A.; Deyev, S.; Stepanov, A.; Gabibov, A. Modular approach to CAR-T regulation based on the barnase-barstar complex for therapy of oncological diseases. In FEBS Open Bio; WILEY: Hoboken, NJ, USA, 2021; Volume 11, p. 93.
- Zdobnova, T.A.; Stremovskiy, O.A.; Lebedenko, E.N.; Deyev, S.M. Self-Assembling Complexes of Quantum Dots and scFv Antibodies for Cancer Cell Targeting and Imaging. PLoS ONE 2012, 7, e48248.
- Lebedenko, E.N.; Balandin, T.G.; Edelweiss, E.F.; Georgiev, O.; Moiseeva, E.S.; Petrov, R.V.; Deyev, S.M. Visualization of cancer cells by means of the fluorescent EGFP-barnase protein. Dokl. Biochem. Biophys. 2007, 414, 120–123.
- Sreenivasan, V.K.A.; Ivukina, E.A.; Deng, W.; Kelf, T.A.; Zdobnova, T.A.; Lukash, S.V.; Veryugin, B.V.; Stremovskiy, O.A.; Zvyagin, A.V.; Deyev, S.M. Barstar:barnase—A versatile platform for colloidal diamond bioconjugation. J. Mater. Chem. 2011, 21, 65–68.
- Grebenik, E.A.; Nadort, A.; Generalova, A.N.; Nechaev, A.V.; Sreenivasan, V.K.A.; Khaydukov, E.V.; Semchishen, V.A.; Popov, A.P.; Sokolov, V.I.; Akhmanov, A.S.; et al. Feasibility study of the optical imaging of a breast cancer lesion labeled with upconversion nanoparticle biocomplexes. J. Biomed. Opt. 2013, 18, 076004.
- Kotelnikova, P.A.; Shipunova, V.O.; Aghayeva, U.F.; Stremovskiy, O.A.; Nikitin, M.P.; Novikov, I.A.; Schulga, A.A.; Deyev, S.M.; Petrov, R.V. Synthesis of Magnetic Nanoparticles Stabilized by Magnetite-Binding Protein for Targeted Delivery to Cancer Cells. Dokl. Biochem. Biophys. 2018, 481, 198–200.
- Shipunova, V.O.; Zelepukin, I.V.; Stremovskiy, O.A.; Nikitin, M.P.; Care, A.; Sunna, A.; Zvyagin, A.V.; Deyev, S.M. Versatile Platform for Nanoparticle Surface Bioengineering Based on SiO2-Binding Peptide and Proteinaceous Barnase*Barstar Interface. ACS Appl. Mater. Interfaces 2018, 10, 17437–17447.
- Balalaeva, I.V.; Zdobnova, T.A.; Sokolova, E.A.; Deyev, S.M. Targeted delivery of quantum dots to the HER2-expressing tumor using recombinant antibodies. Russ. J. Bioorg. Chem. 2015, 41, 536–542.
- Nikitin, M.P.; Zdobnova, T.A.; Lukash, S.V.; Stremovskiy, O.A.; Deyev, S.M. Protein-assisted self-assembly of multifunctional nanoparticles. Proc. Natl. Acad. Sci. USA 2010, 107, 5827–5832.
- Makarov, A.A.; Kolchinsky, A.; Ilinskaya, O.N. Binase and other microbial RNases as potential anticancer agents. Bioessays 2008, 30, 781–790.
- Fang, E.F.; Ng, T.B. Ribonucleases of different origins with a wide spectrum of medicinal applications. Biochim. Biophys. Acta 2011, 1815, 65–74.
- Ulyanova, V.; Vershinina, V.; Ilinskaya, O. Barnase and binase: Twins with distinct fates. FEBS J. 2011, 278, 3633–3643.
