Trifolium L. is an economically important genus that is characterized by variable karyotypes relating to its ploidy level and basic chromosome numbers. The advent of genomic resources combined with molecular cytogenetics provides an opportunity to develop our understanding of plant genomes in general.
- Trifolium
- Chromosome
1. Introduction
The Fabaceae family (Leguminosae, legume or bean family) is the third-largest flowering plant family, after the Asteraceae and Orchidaceae families [1][2]. It is agronomically important, as it can form a symbiotic association with nitrogen-fixing bacteria. Several species from this family serve as genetic model organisms (e.g., Medicago truncatula Gaertn. , Pisum sativum L., and Lotus japonicus L.). With more than 250 species, the clover genus, Trifolium , is one of the largest genera in this family [1][2][3]. This herbaceous genus acquired its name in reference to the characteristic form of the leaf, usually consisting of three leaflets (trifoliolate), and includes both annual and perennial species occurring natively across a large range of biotopes from meadows and open woodlands to semi-deserts and mountain ridges in temperate and, to a lesser extent, subtropical regions. The genus’s origin has been estimated to have occurred in the Early Miocene, 16–23 million years ago, and its center of origin was first assumed to be in California with its subsequent spread into Asia and hence to Europe and Africa [4]. Later, a new hypothesis was proposed of clovers originating in the Mediterranean region due to their species diversity, including the diversity in their chromosome numbers, and because the greatest occurrence of their endemic species is found in this area, with a secondary center of distribution in North America and East Africa [3][5][6][7]. By contrast, native clovers are absent from Australia and Southeast Asia.
Attempts have been made to divide this genus into natural groups. In the 19th century, Bossier [8] divided the genus into seven sections. A century later, eight subgenera were recognized and revised [1][9]. Better insight into phylogeny and the origin of the genus was facilitated by molecular analyses. These showed that Trifolium is a member of a large clade of legumes that lack one copy of the chloroplast inverted repeat [10][11], and a further molecular phylogenetic analysis of the internal transcribed spacer (ITS) and chloroplast genes provided evidence that most of these proposed sections are not monophyletic [12][13]. The most recent subgeneric classification, based on phylogenetic analyses of 218 species’ ribosomal ITS and chloroplast trnL intron sequences, was proposed by Ellison et al. [3], who divided the genus into two subgenera, Chronosemium and Trifolium , with the further subdivision of Trifolium into eight sections— Glycyrrhizum (2 species), Paramesus (2 species), Lupinaster (3 species), Trifolium (73 species), Trichocephalum (9 species), Vesicastrum (54 species), Trifoliastrum (20 species), and Involucrarium (72 species). In 2014, Trifolium phylogenetic analyses were conducted, based on highly unusual Trifolium plastomes [14].
The economic importance of this genus lies in its agricultural utilization. Historically, clovers, and especially red clovers, have been cultivated in rotation with other crops to maintain soil fertility due to their ability to establish a mutualistic relationship with root-nodulating and nitrogen-fixing bacteria. Their value was later diminished by the advent of nitrogen fertilizers, but the global need for sustainable and conservation agriculture is bringing this historical approach back into focus. Nowadays, many Trifolium species are extensively cultivated as fodder plants ( Trifolium pratense L., Trifolium repens L., Trifolium hybridum L., and Trifolium resupinatum L.), and also as green manure crops to enhance soil fertility and sustainability [15]. Further knowledge about the genomes of both wild and cultivated clovers and an understanding of their evolution will prove to be of great benefit in the future of clover breeding.
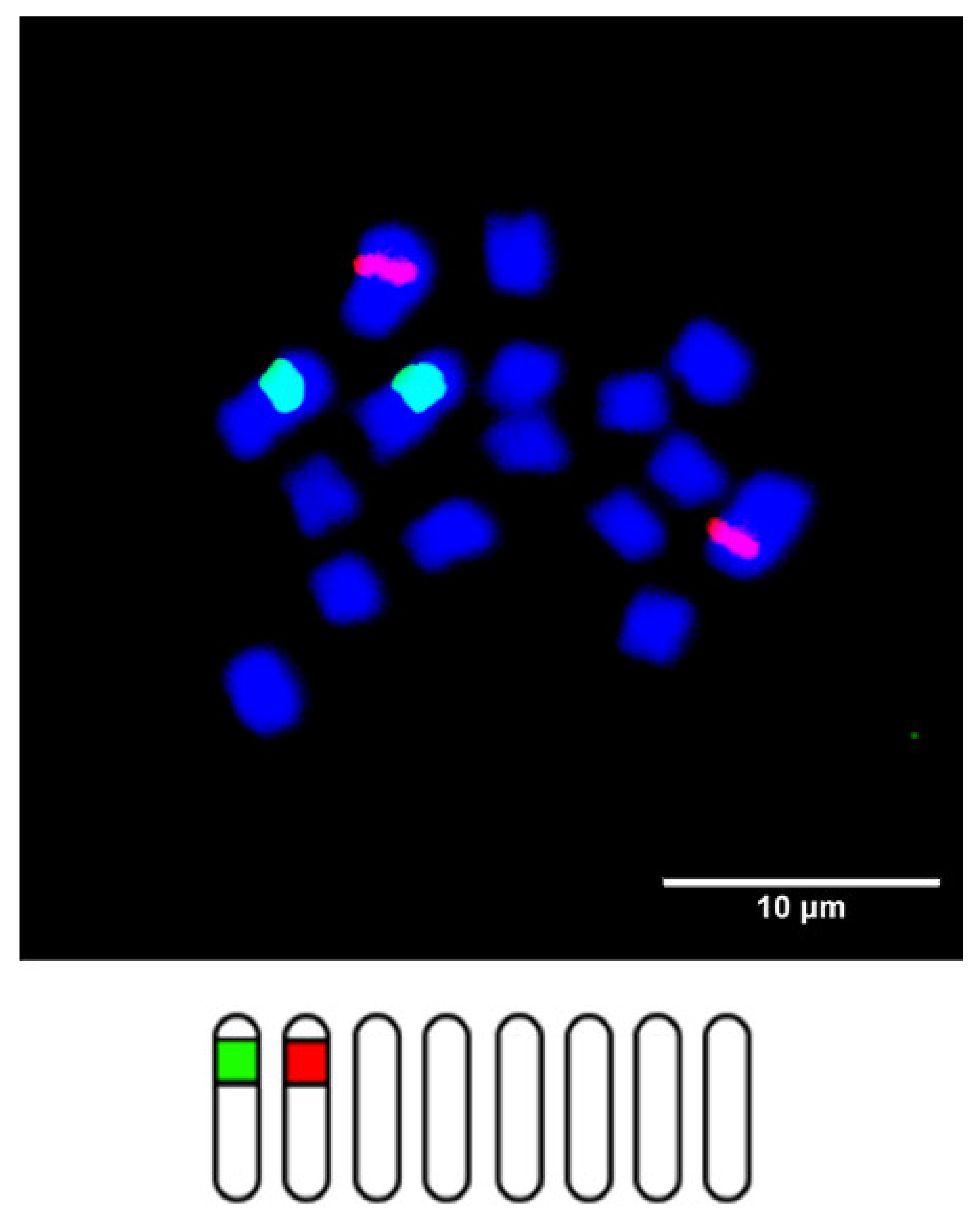
2. Chromosome Identification in Trifolium
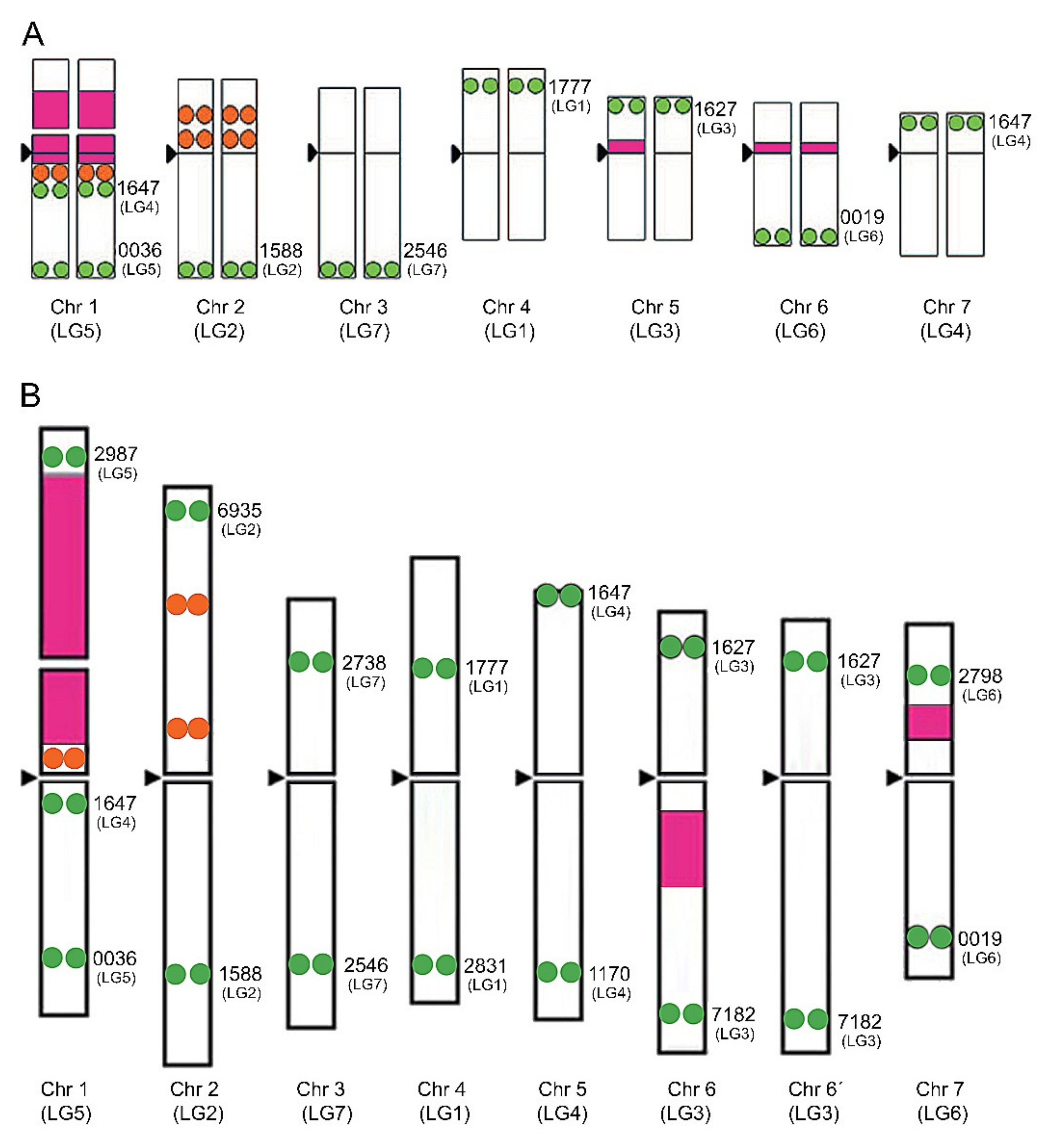
3. Chromosomal Distribution of Ribosomal DNA Genes
| Subgenus/Section | Trifolium Species | 2n | Loci Number per 2n | Reported in | |
|---|---|---|---|---|---|
| 5S | 25S | ||||
| CHRONOSEMIUM | |||||
| T. aureum | 2x = 16 | 4 | 2 | Vozárová et al. [31] | |
| 2x = 14 | 4 | 2 | |||
| T. badium | 2x = 14 | 2 | 4 | ||
| 2 | 2 | ||||
| T. campestre | 2x = 14 | 2 | 2 | Ansari et al. [32] | |
| T. micranthum | 2x = 16 | 2 | 2 | ||
| T. dubium | 4x = 30 | 4 | 4 | ||
| TRIFOLIUM | |||||
| TRIFOLIUM | T. alpestre | 2x = 16 | 10 | 2 | Vozárová et al. [31] |
| 11 | 2 | ||||
| T. arvense | 2x = 14 | 2 | 2 | ||
| T. bocconei | 2x = 12 | 2 | 2 | ||
| T. cherleri | 2x = 10 | 4 | 10 | ||
| T. diffusum | 2x = 16 | 2 | 2 | ||
| T. hirtum | 2x = 10 | 6 | 2 | ||
| T. ligusticum | 2x = 12 | 2 | 2 | ||
| 2x = 14 | 2 | 2 | |||
| T. pallidum | 2x = 16 | 4 | 2 | ||
| T. purpureum | 2x = 14 | 2 | 2 | ||
| T. rubens | 2x = 16 | 4 | 2 | ||
| T. squamosum | 2x = 16 | 4 | 2 | ||
| T. stellatum | 2x = 12 | 4 | 2 | ||
| 2x = 14 | 4 (2w) | 2 | |||
| T. pannonicum | 16x = 128 | 16 | 16 | ||
| T. pratense | 4x = 28 | 8 | 8 | Dluhošová et al. [33] | |
| 2x = 14 | 4 | 5 | Sato et al. [18] | ||
| T. medium | 8x = 64 | 12 | 8 | Dluhošová et al. [33] | |
| TRICHOCEPHALUM | T. subterraneum subsp. subterraneum | 2x = 16 | 2 | 4 (2w) | Vozárová et al. [31] |
| T. subterraneumsubsp. subterraneum | 2x = 16 | 2 | 2 | Falistocco et al. [34] | |
| T. subterraneum subsp. brachycalycinum | 2x = 16 | 2 | 4 (2w) | ||
| T. israeliticum | 2x = 12 | 10 | 4 | ||
| VESICASTRUM | T. fragiferum | 2x = 16 | 2 | 2 | Vozárová et al. [31] |
| T. resupinatum | 2x = 16 | 2 | 2 | ||
| 2x = 14 | 2 | 2 | |||
| T. spumosum | 2x = 16 | 2 | 2 | ||
| 4 | 2 | ||||
| TRIFOLIASTRUM | T. glomeratum | 2x = 16 | 2 | 2 | Vozárová et al. [31] |
| T. montanum | 2x = 16 | 2 | 2 | ||
| T. occidentale | 2x = 16 | 4 | 2 | Ansari [35] | |
| T. pallescens | 2x = 16 | 2 | 2 | Vozárová et al. [31] | |
| T. thalii | 2x = 16 | 2 | 2 | ||
| T. repens | 4x = 32 | 4 | 2 | Ansari [35] | |
| T. uniflorum | 4x = 32 | 4 | 4 | ||
| T. nigrescens subsp. nigrescens | 2x = 16 | 2 | 2 | ||
| T. nigrescens subsp. petrisavii | 2x = 16 | 2 | 2 | ||
| T. ambiguum | 2x = 16 | 2 | 2 | ||
| T. hybridum | 2x = 16 | 2 | 2 | ||
| T. isthmocarpum | 2x = 16 | 2 | 6 | ||
| INVOLUCRARIUM | T. chilense | 2x = 16 | 4 | 2 | Vozárová et al. [31] |
| T. microdon | 2x = 16 | 2 | 2 | ||
| T. microcephalum | 2x = 16 | 16 | 16 | ||
| 16 | 2 | ||||
| PARAMESUS | T. glanduliferum | 2x = 16 | 4 | 2 | Vozárová et al. [31] |
| 5 (1w) | 2 | ||||
| 4 | 2 | ||||
| T. strictum | 2x= 14 | 2 | 2 | ||
| LUPINASTER | T. lupinaster | 4x = 28 | 8 | 4 | Vozárová et al. [31] |
| 4x = 32 | 8 | 4 | |||
4. Conclusions and Future Prospects
References
- Zohary, M.; Heller, D. The Genus Trifolium, 1st ed.; Israel Academy of Sciences and Humanities: Jerusalem, Israel, 1984; pp. 1–610.
- Gillet, J.M.; Taylor, N.L. The World of Clovers, 1st ed.; Iowa State University Press: Ames, IA, USA, 2001; pp. 1–457.
- Ellison, N.W.; Liston, A.; Steiner, J.J.; Williams, W.M.; Taylor, N.L. Molecular phylogenetics of the clover genus (Trifolium-Leguminosae). Mol. Phylogenet. Evol. 2006, 39, 688–705.
- Zohary, M. Origin and evolution in the genus Trifolium. Bot. Notiser. 1972, 125, 501–511.
- Taylor, N.L. Clovers Around the World. In Agronomy Monographs; Taylor, N.L., Ed.; Soil Science Society of America: Madison, WI, USA, 1985; Volume 25, pp. 1–6.
- Panitsa, M.; Trigas, P.; Iatrou, G.; Sfenthourakis, S. Factors affecting plant species richness and endemism on land-bridge islands—An example from the East Aegean Archipelago. Acta Oceol. 2010, 36, 431–437.
- Scoppola, A.; Tirado, J.L.; Gutiérrez, F.M.; Magrini, S. The genus Trifolium (Fabaceae) in South Europe: A critical review on species richness and distribution. Nord. J. Bot. 2018, 36, e01723.
- Boissier, E. Trifolium. In Flora Orientalis; H. Georg: Basileae, Switzerland, 1872; pp. 110–156.
- Hossain, M. A revision of Trifolium in the nearer East. Notes R. Bot. Gard. Edinb. 1961, 23, 387–481.
- Lavin, M.; Doyle, J.J.; Palmer, J.D. Evolutionary significance of the loss of the chloroplast-DNA inverted repeat in the Leguminosae subfamily Papilionoideae. Evolution 1990, 44, 390–402.
- Liston, A. Use of the polymerase chain reaction to survey for the loss of the inverted repeat in the legume chloroplast genome. In Advances in Legume Systematics; Crisp, M.D., Doyle, J.J., Eds.; Royal Botanic Gardens: Kew, UK, 1995; Volume 7, pp. 31–40.
- Watson, L.E.; Sayed-Ahmed, H.; Badr, A. Molecular phylogeny of Old World Trifolium (Fabaceae). Plant Syst. Evol. 2000, 224, 153–171.
- Steele, K.; Wojciechowski, M. Phylogenetic analyses of tribes Trifolieae and Vicieae, based on sequences of the plastid gene matK (Papilionoideae: Leguminosae). Adv. Legume Syst. 2003, 1, 355–370.
- Sveinsson, S.; Cronk, Q. Evolutionary origin of highly repetitive plastid genomes within the clover genus (Trifolium). BMC Evol. Biol. 2014, 14, 228.
- Kintl, A.; Elbl, J.; Lošák, T.; Vaverková, M.; Nedělník, J. Mixed intercropping of wheat and white clover to enhance the sustainability of the conventional cropping system: Effects on biomass production and leaching of mineral nitrogen. Sustainability 2018, 10, 3367.
- Fuchs, J.; Strehl, S.; Brandes, A.; Schweizer, D.; Schubert, I. Molecular-cytogenetic characterization of the Vicia faba genome—Heterochromatin differentiation, replication patterns and sequence localization. Chromosome Res. 1998, 6, 219–230.
- Fuchs, J.; Kühne, M.; Schubert, I. Assignment of linkage groups to pea chromosomes after karyotyping and gene mapping by fluorescent in situ hybridization. Chromosoma 1998, 107, 272–276.
- Sato, S.; Isobe, S.; Asamizu, E.; Ohmido, N.; Kataoka, R.; Nakamura, Y.; Kaneko, T.; Sakurai, N.; Okumura, K.; Klimenko, I.; et al. Comprehensive structural analysis of the genome of red clover (Trifolium pratense L.). DNA Res. 2005, 12, 301–364.
- Kataoka, R.; Hara, M.; Kato, S.; Isobe, S.; Sato, S.; Tabata, S.; Ohmido, N. Integration of linkage and chromosome maps of red clover (Trifolium pratense L.). Cytogenet. Genome Res. 2012, 137, 60–69.
- De Oliveira Bustamante, F.; do Nascimento, T.H.; Montenegro, C.; Dias, S.; do Vale Martins, L.; Braz, G.T.; Benko-Iseppon, A.M.; Jiang, J.; Pedrosa-Harand, A.; Brasileiro-Vidal, A.C. Oligo-FISH barcode in beans: A new chromosome identification system. Theor. Appl. Genet. 2021, 134, 3675–3686.
- Montenegro, C.; do Vale Martins, L.; de Oliveira Bustamante, F.; Brasileiro-Vidal, A.C.; Pedrosa-Harand, A. Comparative cytogenomics reveals genome reshuffling and centromere repositioning in the legume tribe Phaseoleae. bioRxiv 2021.
- Biscotti, M.A.; Olmo, E.; Heslop-Harrison, J.S. Repetitive DNA in eukaryotic genomes. Chromosome Res. 2015, 23, 415–420.
- Flavell, R.B. The structure and control of expression of ribosomal RNA genes. Oxf. Surv. Plant Mol. Cell Biol. 1986, 3, 251–274.
- Dvořák, J.; Zhang, H.-B.; Kota, R.S.; Lassner, M. Organization and evolution of the 5S ribosomal rna gene family in wheat and related species. Genome 1989, 32, 1003–1016.
- Schubert, I.; Wobus, U. In situ hybridization confirms jumping nucleolus organizing regions in Allium. Chromosoma 1985, 92, 143–148.
- Raina, S.N.; Mukai, Y. Detection of a variable number of 18S-5.8S-26S and 5S ribosomal DNA loci by fluorescent in situ hybridization in diploid and tetraploid Arachis species. Genome 1999, 42, 52–59.
- Pedrosa-Harand, A.; de Almeida, C.C.S.; Mosiolek, M.; Blair, M.W.; Schweizer, D.; Guerra, M. Extensive ribosomal DNA amplification during andean common bean (Phaseolus vulgaris L.) evolution. Theor. Appl. Genet. 2006, 112, 924–933.
- Chung, M.-C.; Lee, Y.-I.; Cheng, Y.-Y.; Chou, Y.-J.; Lu, C.-F. Chromosomal polymorphism of ribosomal genes in the genus Oryza. Theor. Appl. Genet. 2008, 116, 745–753.
- Roa, F.; Guerra, M. Distribution of 45S rDNA sites in chromosomes of plants: Structural and evolutionary implications. BMC Evol. Biol. 2012, 12, 225.
- Roa, F.; Guerra, M. Non-Random Distribution of 5S rDNA sites and its association with 45S rDNA in plant chromosomes. Cytogenet. Genome Res. 2015, 146, 243–249.
- Vozárová, R.; Macková, E.; Vlk, D.; Řepková, J. Variation in ribosomal DNA in the genus Trifolium (Fabaceae). Plants 2021, 10, 1771.
- Ansari, H.A.; Ellison, N.W.; Williams, W.M. Molecular and cytogenetic evidence for an allotetraploid origin of Trifolium dubium (Leguminosae). Chromosoma 2008, 117, 159–167.
- Dluhošová, J.; Řepková, J.; Jakešová, H.; Nedělník, J. Impact of interspecific hybridization of T. pratense × T. medium and backcrossing on genetic variability of progeny. Czech J. Genet. Plant 2016, 52, 125–131.
- Falistocco, E.; Marconi, G.; Falcinelli, M. Comparative cytogenetic study on Trifolium subterraneum (2n = 16) and Trifolium israeliticum (2n = 12). Genome 2013, 56, 307–313.
- Ansari, H. Molecular cytogenetic organization of 5S and 18S-26S rDNA loci in white clover (Trifolium repens L.) and related species. Ann. Bot. Lond. 1999, 83, 199–206.
- Jiang, J.; Gill, B.S.; Wang, G.L.; Ronald, P.C.; Ward, D.C. Metaphase and interphase fluorescence in situ hybridization mapping of the rice genome with bacterial artificial chromosomes. Proc. Natl. Acad. Sci. USA 1995, 92, 4487–4491.
- Lapitan, N.L.V.; Brown, S.E.; Kennard, W.; Stephens, J.L.; Knudson, D.L. FISH physical mapping with barley BAC clones. Plant J. 1997, 11, 149–156.
- Peterson, D.G.; Lapitan, N.L.; Stack, S.M. Localization of single- and low-copy sequences on tomato synaptonemal complex spreads using fluorescence in situ hybridization (FISH). Genetics 1999, 152, 427–439.
- Zhong, X.-B.; Bodeau, J.; Fransz, P.F.; Williamson, V.M.; van Kammen, A.; de Jong, J.H.; Zabel, P. FISH to meiotic pachytene chromosomes of tomato locates the root-knot nematode resistance gene Mi-1 and the acid phosphatase gene Aps-1 near the junction of euchromatin and pericentromeric heterochromatin of chromosome arms 6S and 6L, respectively: Theor. Appl. Genet. 1999, 98, 365–370.
- Islam-Faridi, M.N.; Childs, K.L.; Klein, P.E.; Hodnett, G.; Menz, M.A.; Klein, R.R.; Rooney, W.L.; Mullet, J.E.; Stelly, D.M.; Price, H.J. A molecular cytogenetic map of sorghum Chromosome 1. Fluorescence in situ hybridization analysis with mapped bacterial artificial chromosomes. Genetics 2002, 161, 345–353.
- Lee, H.-R.; Eom, E.-M.; Lim, Y.-P.; Bang, J.-W.; Lee, D.-H. Construction of a garlic BAC library and chromosomal assignment of BAC clones using the FISH technique. Genome 2003, 46, 514–520.
- Schnabel, E.; Kulikova, O.; Penmetsa, R.V.; Bisseling, T.; Cook, D.R.; Frugoli, J. An integrated physical, genetic and cytogenetic map around the sunn locus of Medicago truncatula. Genome 2003, 46, 665–672.
- Bonifácio, E.M.; Fonsêca, A.; Almeida, C.; Dos Santos, K.G.B.; Pedrosa-Harand, A. Comparative cytogenetic mapping between the lima bean (Phaseolus lunatus L.) and the common bean (P. vulgaris L.). Theor. Appl. Genet. 2012, 124, 1513–1520.
- Lysak, M.A.; Fransz, P.F.; Ali, H.B.; Schubert, I. Chromosome painting in Arabidopsis thaliana. Plant J. 2001, 28, 689–697.
- Pecinka, A.; Schubert, V.; Meister, A.; Kreth, G.; Klatte, M.; Lysak, M.A.; Fuchs, J.; Schubert, I. Chromosome territory arrangement and homologous pairing in nuclei of Arabidopsis thaliana are predominantly random except for NOR-bearing chromosomes. Chromosoma 2004, 113, 258–269.
- Betekhtin, A.; Jenkins, G.; Hasterok, R. Reconstructing the evolution of Brachypodium genomes using comparative chromosome painting. PLoS ONE 2014, 9, e115108.
- Han, Y.; Zhang, T.; Thammapichai, P.; Weng, Y.; Jiang, J. Chromosome-specific painting in Cucumis species using bulked oligonucleotides. Genetics 2015, 200, 771–779.
- Braz, G.T.; He, L.; Zhao, H.; Zhang, T.; Semrau, K.; Rouillard, J.-M.; Torres, G.A.; Jiang, J. Comparative oligo-FISH mapping: An efficient and powerful methodology to reveal karyotypic and chromosomal evolution. Genetics 2018, 208, 513–523.
- Qu, M.; Li, K.; Han, Y.; Chen, L.; Li, Z.; Han, Y. Integrated karyotyping of woodland strawberry (Fragaria vesca) with oligopaint FISH probes. Cytogenet. Genome Res. 2017, 153, 158–164.
- Albert, P.S.; Zhang, T.; Semrau, K.; Rouillard, J.-M.; Kao, Y.-H.; Wang, C.-J.R.; Danilova, T.V.; Jiang, J.; Birchler, J.A. Whole-chromosome paints in maize reveal rearrangements, nuclear domains, and chromosomal relationships. Proc. Natl. Acad. Sci. USA 2019, 116, 1679–1685.
- Liu, X.; Sun, S.; Wu, Y.; Zhou, Y.; Gu, S.; Yu, H.; Yi, C.; Gu, M.; Jiang, J.; Liu, B.; et al. Dual-color oligo-FISH can reveal chromosomal variations and evolution in Oryza species. Plant J. 2020, 101, 112–121.
- Šimoníková, D.; Němečková, A.; Karafiátová, M.; Uwimana, B.; Swennen, R.; Doležel, J.; Hřibová, E. Chromosome painting facilitates anchoring reference genome sequence to chromosomes in situ and integrated karyotyping in banana (Musa spp.). Front. Plant Sci. 2019, 10, 1503.
- Li, G.; Zhang, T.; Yu, Z.; Wang, H.; Yang, E.; Yang, Z. An efficient oligo-FISH painting system for revealing chromosome rearrangements and polyploidization in Triticeae. Plant J. 2021, 105, 978–993.
- Do Vale Martins, L.; de Oliveira Bustamante, F.; da Silva Oliveira, A.R.; da Costa, A.F.; de Lima Feitoza, L.; Liang, Q.; Zhao, H.; Benko-Iseppon, A.M.; Muñoz-Amatriaín, M.; Pedrosa-Harand, A.; et al. BAC- and oligo-FISH mapping reveals chromosome evolution among Vigna angularis, V. unguiculata, and Phaseolus vulgaris. Chromosoma 2021, 130, 133–147.
