Trifolium L. is an economically important genus that is characterized by variable karyotypes relating to its ploidy level and basic chromosome numbers. The advent of genomic resources combined with molecular cytogenetics provides an opportunity to develop our understanding of plant genomes in general.
- Trifolium
- Chromosome
1. Introduction
The Fabaceae family (Leguminosae, legume or bean family) is the third-largest flowering plant family, after the Asteraceae and Orchidaceae families [1][2][1,2]. It is agronomically important, as it can form a symbiotic association with nitrogen-fixing bacteria. Several species from this family serve as genetic model organisms (e.g., Medicago truncatula Gaertn. , Pisum sativum L., and Lotus japonicus L.). With more than 250 species, the clover genus, Trifolium , is one of the largest genera in this family [1][2][3][1,2,3]. This herbaceous genus acquired its name in reference to the characteristic form of the leaf, usually consisting of three leaflets (trifoliolate), and includes both annual and perennial species occurring natively across a large range of biotopes from meadows and open woodlands to semi-deserts and mountain ridges in temperate and, to a lesser extent, subtropical regions. The genus’s origin has been estimated to have occurred in the Early Miocene, 16–23 million years ago, and its center of origin was first assumed to be in California with its subsequent spread into Asia and hence to Europe and Africa [4]. Later, a new hypothesis was proposed of clovers originating in the Mediterranean region due to their species diversity, including the diversity in their chromosome numbers, and because the greatest occurrence of their endemic species is found in this area, with a secondary center of distribution in North America and East Africa [3][5][6][7][3,5,6,7]. By contrast, native clovers are absent from Australia and Southeast Asia.
Attempts have been made to divide this genus into natural groups. In the 19th century, Bossier [8] divided the genus into seven sections. A century later, eight subgenera were recognized and revised [1][9][1,9]. Better insight into phylogeny and the origin of the genus was facilitated by molecular analyses. These showed that Trifolium is a member of a large clade of legumes that lack one copy of the chloroplast inverted repeat [10][11][10,11], and a further molecular phylogenetic analysis of the internal transcribed spacer (ITS) and chloroplast genes provided evidence that most of these proposed sections are not monophyletic [12][13][12,13]. The most recent subgeneric classification, based on phylogenetic analyses of 218 species’ ribosomal ITS and chloroplast trnL intron sequences, was proposed by Ellison et al. [3], who divided the genus into two subgenera, Chronosemium and Trifolium , with the further subdivision of Trifolium into eight sections— Glycyrrhizum (2 species), Paramesus (2 species), Lupinaster (3 species), Trifolium (73 species), Trichocephalum (9 species), Vesicastrum (54 species), Trifoliastrum (20 species), and Involucrarium (72 species). In 2014, Trifolium phylogenetic analyses were conducted, based on highly unusual Trifolium plastomes [14].
The economic importance of this genus lies in its agricultural utilization. Historically, clovers, and especially red clovers, have been cultivated in rotation with other crops to maintain soil fertility due to their ability to establish a mutualistic relationship with root-nodulating and nitrogen-fixing bacteria. Their value was later diminished by the advent of nitrogen fertilizers, but the global need for sustainable and conservation agriculture is bringing this historical approach back into focus. Nowadays, many Trifolium species are extensively cultivated as fodder plants ( Trifolium pratense L., Trifolium repens L., Trifolium hybridum L., and Trifolium resupinatum L.), and also as green manure crops to enhance soil fertility and sustainability [15]. Further knowledge about the genomes of both wild and cultivated clovers and an understanding of their evolution will prove to be of great benefit in the future of clover breeding.
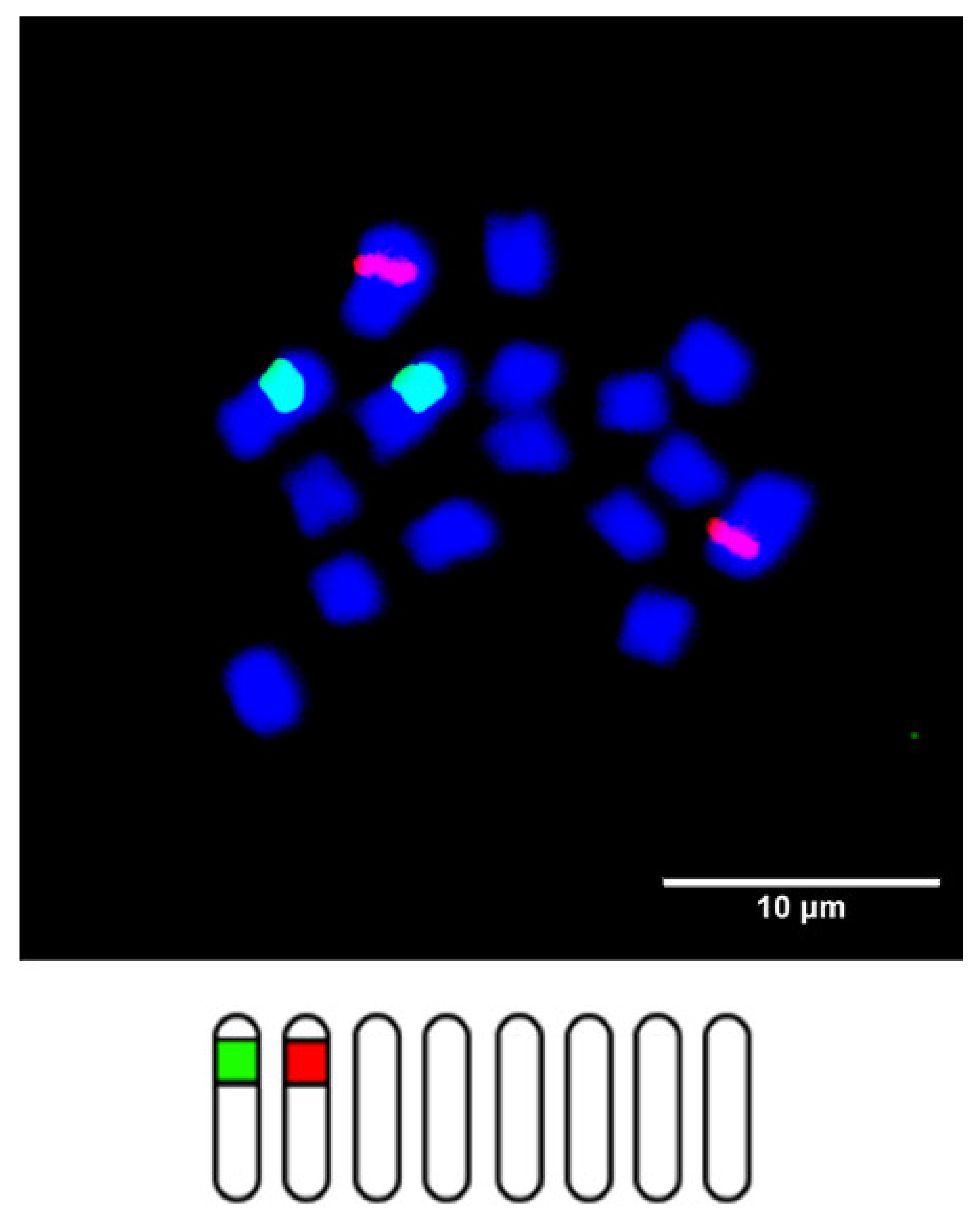
2. Chromosome Identification in Trifolium
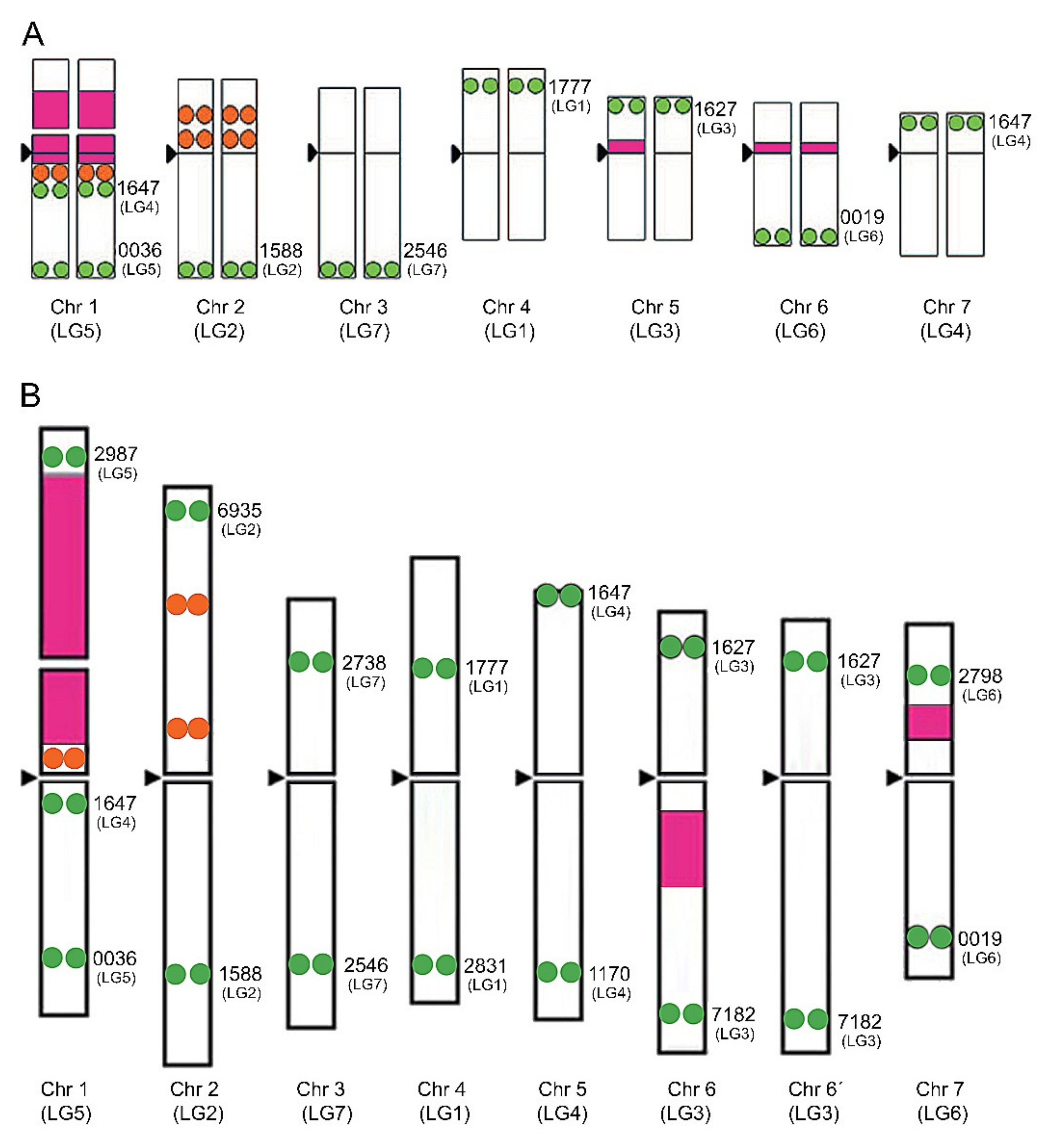
3. Chromosomal Distribution of Ribosomal DNA Genes
| Subgenus/Section | Trifolium | Species | 2n | Loci Number per 2n | Reported in | |
|---|---|---|---|---|---|---|
| 5S | 25S | |||||
| CHRONOSEMIUM | ||||||
| T. aureum | 2x = 16 | 4 | 2 | Vozárová et al. [31] | ||
| 2x = 14 | 4 | 2 | ||||
| T. badium | 2x = 14 | 2 | 4 | |||
| 2 | 2 | |||||
| T. campestre | 2x = 14 | 2 | 2 | Ansari et al. [32] | Ansari et al. [58] | |
| T. micranthum | 2x = 16 | 2 | 2 | |||
| T. dubium | 4x = 30 | 4 | 4 | |||
| TRIFOLIUM | ||||||
| TRIFOLIUM | T. alpestre | 2x = 16 | 10 | 2 | Vozárová et al. [31] | |
| 11 | 2 | |||||
| T. arvense | 2x = 14 | 2 | 2 | |||
| T. bocconei | 2x = 12 | 2 | 2 | |||
| T. cherleri | 2x = 10 | 4 | 10 | |||
| T. diffusum | 2x = 16 | 2 | 2 | |||
| T. hirtum | 2x = 10 | 6 | 2 | |||
| T. ligusticum | 2x = 12 | 2 | 2 | |||
| 2x = 14 | 2 | 2 | ||||
| T. pallidum | 2x = 16 | 4 | 2 | |||
| T. purpureum | 2x = 14 | 2 | 2 | |||
| T. rubens | 2x = 16 | 4 | 2 | |||
| T. squamosum | 2x = 16 | 4 | 2 | |||
| T. stellatum | 2x = 12 | 4 | 2 | |||
| 2x = 14 | 4 (2w) | 2 | ||||
| T. pannonicum | 16x = 128 | 16 | 16 | |||
| T. pratense | 4x = 28 | 8 | 8 | Dluhošová et al. [33] | Dluhošová et al. [80] | |
| 2x = 14 | 4 | 5 | Sato et al. [18] | Sato et al. [43] | ||
| T. medium | 8x = 64 | 12 | 8 | Dluhošová et al. [33] | Dluhošová et al. [80] | |
| TRICHOCEPHALUM | T. subterraneum subsp. subterraneum | 2x = 16 | 2 | 4 (2w) | Vozárová et al. [31] | |
| T. subterraneumsubsp. subterraneum | 2x = 16 | 2 | 2 | Falistocco et al. [34] | Falistocco et al. [81] | |
| T. subterraneum subsp. brachycalycinum | 2x = 16 | 2 | 4 (2w) | |||
| T. israeliticum | 2x = 12 | 10 | 4 | |||
| VESICASTRUM | T. fragiferum | 2x = 16 | 2 | 2 | Vozárová et al. [31] | |
| T. resupinatum | 2x = 16 | 2 | 2 | |||
| 2x = 14 | 2 | 2 | ||||
| T. spumosum | 2x = 16 | 2 | 2 | |||
| 4 | 2 | |||||
| TRIFOLIASTRUM | T. glomeratum | 2x = 16 | 2 | 2 | Vozárová et al. [31] | |
| T. montanum | 2x = 16 | 2 | 2 | |||
| T. occidentale | 2x = 16 | 4 | 2 | Ansari [35] | Ansari [82] | |
| T. pallescens | 2x = 16 | 2 | 2 | Vozárová et al. [31] | ||
| T. thalii | 2x = 16 | 2 | 2 | |||
| T. repens | 4x = 32 | 4 | 2 | Ansari [35] | Ansari [82] | |
| T. uniflorum | 4x = 32 | 4 | 4 | |||
| T. nigrescens subsp. nigrescens | 2x = 16 | 2 | 2 | |||
| T. nigrescens subsp. petrisavii | 2x = 16 | 2 | 2 | |||
| T. ambiguum | 2x = 16 | 2 | 2 | |||
| T. hybridum | 2x = 16 | 2 | 2 | |||
| T. isthmocarpum | 2x = 16 | 2 | 6 | |||
| INVOLUCRARIUM | T. chilense | 2x = 16 | 4 | 2 | Vozárová et al. [31] | |
| T. microdon | 2x = 16 | 2 | 2 | |||
| T. microcephalum | 2x = 16 | 16 | 16 | |||
| 16 | 2 | |||||
| PARAMESUS | T. glanduliferum | 2x = 16 | 4 | 2 | Vozárová et al. [31] | |
| 5 (1w) | 2 | |||||
| 4 | 2 | |||||
| T. strictum | 2x= 14 | 2 | 2 | |||
| LUPINASTER | T. lupinaster | 4x = 28 | 8 | 4 | Vozárová et al. [31] | |
| 4x = 32 | 8 | 4 | ||||
