COVID-19 is caused by a coronavirus called severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2). The difficulty in containing SARS-CoV-2 has underscored the need for techniques such as mass spectrometry in the diagnosis and treatment of COVID-19. Mass spectrometry-based methods have been employed in several studies to detect changes in interactions among host proteins, and between host and viral proteins in COVID-19 patients. The methods have also been used to characterize host and viral proteins, and analyze lipid metabolism in COVID-19 patients. Information obtained using the above methods are complemented by high-throughput analysis of transcriptomic and epigenomic changes associated with COVID-19, coupled with next-generation sequencing.
- mass spectrometry
- proteomics
- liquid chromatography
- lipidomics
- inflammation
1. Proteomics and Mass Spectrometry in Coronavirus Disease 2019 (COVID-19)
1.1. COVID-19-Linked Host Protein Characterization Discovered through Mass Spectrometric Proteomics
1.2. Proteomics in COVID-19 Treatment Identification
Several treatments for COVID-19 have been identified through MS-based proteomics; the development of these treatments relies on the identification of SARS-CoV-2 replication machinery. A study by Gordon et al. pursued ligands of SARS-CoV-2 interacting proteins in an attempt to distinguish targets for treatment [9]. After the discovery of 69 such compounds, further testing was performed to name two successful treatment classes: inhibitors of protein synthesis (such as ternatin-4 and zotatifin) and ligands of Sigma receptors (such as hydroxychloroquine and haloperidol). Both classes were observed to have effects on SARS-CoV-2 replication, identified through a decrease in the viral nucleoprotein over an 8 h timespan. The prior identification of virus-host protein interactions through AP-MS was an integral step in the discovery of the two treatment classes [9]. In addition to host–virus protein interactions, the discovery of case severity related protein biomarkers has also contributed to the identification of COVID-19 treatments. By distinguishing 38 proteins with differing levels of expression in mild versus severe cases of COVID-19, utilizing label-free quantitative MS, Suvarna et al. identified disruptions in several cellular processes. These processes include blood coagulation, for which proteins such as alpha-2-macroglobulin and fibrinogen gamma chain are upregulated in severe COVID-19 cases, and inflammation, for which proteins such as Serpin Family A Member 3 are upregulated in severe COVID-19 cases. Nine of the 38 identified proteins were then utilized to test the efficacy of COVID-19 treatments, resulting in additional supporting evidence for previously identified treatments such as Selinexor (an inhibitor of viral replication) [10].2. Lipidomics and Mass Spectrometry in COVID-19
2.1. The Immunomodulatory Axis of COVID-19 Disease Burden
2.3. Mass Spectrometry-Based Studies on Spike Protein Helps in Vaccine Development
2.4. Signaling Pattern-Recognition Receptors (PRRs) Are Found on Multiple Host-Cell Surfaces
2.5. Sphingolipids at the Center of COVID-19 Infection Dynamics
There is good reason to assume the protein transmembrane domains, typically composed of hydrophobic amino acid side chains, induce their insertion into the membrane via higher order lipid domain transformations from helix type II to bilayer transitions as coordinated by surface tension molecular regulation [34]. In a recent review of sphingolipids and SARS-CoV-2, it was established that lung cell ceramide facilitated the infection process, while sphingosine-1P inhibited it [35]. This implicates the sphingomyelinase biosynthetic route since the neutral SMAse isoform of the enzyme is located in the plasma membrane and upon activation would generate in situ CER from membrane sphingomyelin thus promoting this metabolic route for viral entry instead of de novo synthesis from palmitoyl CoA plus L-serine or the salvage routes through sphingosine phosphate or glycosphingolipid pathways. However, acid SMase (aSMase) inhibitors such as amitriptyline and pharmacotherapeutics such as hydroxychloroquine that raise the pH of the phagolysosome (thus inhibiting aSMase) may suggest intracellular CER synthesis and thus membrane lipid raft assembly is also involved in the decrease of COVID-19 transmission and disease severity. Figure 1 provides a general outline for the proteomic/lipidomic axis in COVID-19 transmittance and pathology.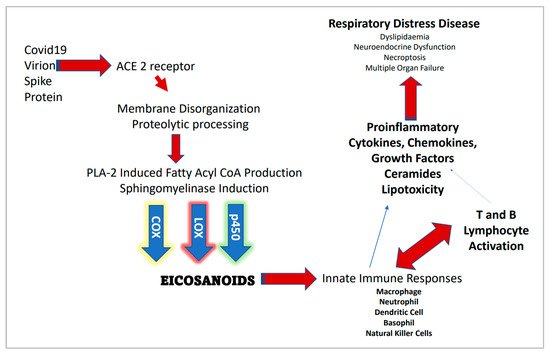
2.6. Contributions of Lipidomics in Understanding and Treating COVID-19
References
- Jannetto, P.J.; Danso, D. Mass spectrometry. Clin. Biochem. 2020, 82, 1.
- Beccaria, M.; Cabooter, D. Current developments in LC-MS for pharmaceutical analysis. Analyst 2020, 145, 1129–1157.
- Seger, C.; Salzmann, L. After another decade: LC–MS/MS became routine in clinical diagnostics. Clin. Biochem. 2020, 82, 2–11.
- Nikolaev, E.N.; Indeykina, M.I.; Brzhozovskiy, A.G.; Bugrova, A.E.; Kononikhin, A.S.; Starodubtseva, N.L.; Petrotchenko, E.V.; Kovalev, G.I.; Borchers, C.H.; Sukhikh, G.T. Mass-Spectrometric Detection of SARS-CoV-2 Virus in Scrapings of the Epithelium of the Nasopharynx of Infected Patients via Nucleocapsid N Protein. J. Proteome Res. 2020, 19, 4393–4397.
- Rybicka, M.; Milosz, E.; Bielawski, K.P. Superiority of MALDI-TOF Mass Spectrometry over Real-Time PCR for SARS-CoV-2 RNA Detection. Viruses 2021, 13, 730.
- Hober, A.; Hua, T.M.K.; Foley, D.; McDonald, T.; Vissers, J.P.; Pattison, R.; Ferries, S.; Hermansson, S.; Betner, I.; Uhlen, M.; et al. Rapid and Sensitive Detection of SARS-CoV-2 Infection Using Quantitative Peptide Enrichment LC-MS/MS Analysis. medRxiv 2021.
- Wang, W.; Li, Q.; Huang, G.; Lin, B.-Y.; Lin, D.-Z.; Ma, Y.; Zhang, Z.; Chen, T.; Zhou, J. Tandem Mass Tag-Based Proteomic Analysis of Potential Biomarkers for Hepatocellular Carcinoma Differentiation. OncoTargets Ther. 2021, 14, 1007–1020.
- Shen, B.; Yi, X.; Sun, Y.; Bi, X.; Du, J.; Zhang, C.; Quan, S.; Zhang, F.; Sun, R.; Qian, L.; et al. Proteomic and Metabolomic Characterization of COVID-19 Patient Sera. Cell 2020, 182, 59–72.e15.
- Gordon, D.E.; Jang, G.M.; Bouhaddou, M.; Xu, J.; Obernier, K.; White, K.M.; O’Meara, M.J.; Rezelj, V.V.; Guo, J.Z.; Swaney, D.L.; et al. A SARS-CoV-2 protein interaction map reveals targets for drug repurposing. Nature 2020, 583, 459–468.
- Suvarna, K.; Biswas, D.; Pai, M.G.J.; Acharjee, A.; Bankar, R.; Palanivel, V.; Salkar, A.; Verma, A.; Mukherjee, A.; Choudhury, M.; et al. Proteomics and Machine Learning Approaches Reveal a Set of Prognostic Markers for COVID-19 Severity With Drug Repurposing Potential. Front. Physiol. 2021, 12, 652799.
- Chilosi, M.; Poletti, V.; Ravaglia, C.; Rossi, G.; Dubini, A.; Piciucchi, S.; Pedica, F.; Bronte, V.; Pizzolo, G.; Martignoni, G.; et al. The pathogenic role of epithelial and endothelial cells in early-phase COVID-19 pneumonia: Victims and partners in crime. Mod. Pathol. 2021, 34, 1444–1455.
- Chua, R.L.; Lukassen, S.; Trump, S.; Hennig, B.P.; Wendisch, D.; Pott, F.; Debnath, O.; Thürmann, L.; Kurth, F.; Völker, M.T.; et al. COVID-19 severity correlates with airway epithelium–immune cell interactions identified by single-cell analysis. Nat. Biotechnol. 2020, 38, 970–979.
- Hammock, B.D.; Wang, W.; Gilligan, M.M.; Panigrahyyz, D. Eicosanoids: The Overlooked Storm in Coronavirus Disease 2019 (COVID-19)? Am. J. Pathol. 2020, 190, 1782–1788.
- Penttilä, P.A.; van Gassen, S.; Panovska, D.; Vanderbeke, L.; van Herck, Y.; Quintelier, K.; Emmaneel, A.; Filtjens, J.; Malengier-Devlies, B.; Ahmadzadeh, K.; et al. High dimensional profiling identifies specific immune types along the recovery trajectories of critically ill COVID19 patients. Experientia 2021, 78, 3987–4002.
- Sichitiu, J.; Fakhouri, F.; Desseauve, D. Antenatal corticosteroid therapy and COVID-19: Pathophysiological considerations. Acta Obstet. Gynecol. Scand. 2020, 99, 952.
- Praissman, J.; Wells, L. Proteomics-Based Insights Into the SARS-CoV-2-Mediated COVID-19 Pandemic: A Review of the First Year of Research. Mol. Cell. Proteom. 2021, 20, 100103.
- Miller, L.M.; Barnes, L.F.; Raab, S.A.; Draper, B.E.; El-Baba, T.J.; Lutomski, C.A.; Robinson, C.V.; Clemmer, D.E.; Jarrold, M.F. Heterogeneity of Glycan Processing on Trimeric SARS-CoV-2 Spike Protein Revealed by Charge Detection Mass Spectrometry. J. Am. Chem. Soc. 2021, 143, 3959–3966.
- Roberts, D.S.; Mann, M.W.; Melby, J.A.; Larson, E.J.; Zhu, Y.; Brasier, A.R.; Jin, S.; Ge, Y. Structural O-Glycoform Heterogeneity of the SARS-CoV-2 Spike Protein Receptor-Binding Domain Revealed by Top-Down Mass Spectrometry. J. Am. Chem. Soc. 2021, 143, 12014–12024.
- Cho, B.G.; Gautam, S.; Peng, W.; Huang, Y.; Goli, M.; Mechref, Y. Direct Comparison of N-Glycans and Their Isomers Derived from Spike Glycoprotein 1 of MERS-CoV, SARS-CoV-1, and SARS-CoV-2. J. Proteome Res. 2021, 20, 4357–4365.
- Antonopoulos, A.; Broome, S.; Sharov, V.; Ziegenfuss, C.; Easton, R.L.; Panico, M.; Dell, A.; Morris, H.R.; Haslam, S.M. Site-specific characterization of SARS-CoV-2 spike glycoprotein receptor-binding domain. Glycobiology 2021, 31, 181–187.
- Witkowska, D. Mass Spectrometry and Structural Biology Techniques in the Studies on the Coronavirus-Receptor Interaction. Molecules 2020, 25, 4133.
- Krishnan, S.; Krishnan, G.P. N-Glycosylation Network Construction and Analysis to Modify Glycans on the Spike (S) Glycoprotein of SARS-CoV-2. Front. Bioinform. 2021, 1.
- Watanabe, Y.; Berndsen, Z.T.; Raghwani, J.; Seabright, G.E.; Allen, J.D.; Pybus, O.; McLellan, J.S.; Wilson, I.A.; Bowden, T.A.; Ward, A.B.; et al. Vulnerabilities in coronavirus glycan shields despite extensive glycosylation. Nat. Commun. 2020, 11, 2688.
- Casalino, L.; Gaieb, Z.; Goldsmith, J.A.; Hjorth, C.K.; Dommer, A.C.; Harbison, A.M.; Fogarty, C.A.; Barros, E.P.; Taylor, B.C.; McLellan, J.S.; et al. Beyond Shielding: The Roles of Glycans in the SARS-CoV-2 Spike Protein. ACS Cent. Sci. 2020, 6, 1722–1734.
- Petrović, T.; Alves, I.; Bugada, D.; Pascual, J.; Vučković, F.; Skelin, A.; Gaifem, J.; Villar-Garcia, J.; Vicente, M.M.; Fernandes, Â.; et al. Composition of the immunoglobulin G glycome associates with the severity of COVID-19. Glycobiology 2020, 31, 372–377.
- Trkulja, V.; Kodvanj, I.; Homolak, J. Immunoglobulin G glycome and severity of COVID-19: More likely a quantification of bias than a true association. A comment on Petrović et al., “Composition of the immunoglobulin G glycome associates with the severity of COVID-19”. Glycobiology 2021, 31, 713–716.
- Hou, H.; Yang, H.; Liu, P.; Huang, C.; Wang, M.; Li, Y.; Zhu, M.; Wang, J.; Xu, Y.; Wang, Y.; et al. Profile of Immunoglobulin G N-Glycome in COVID-19 Patients: A Case-Control Study. Front. Immunol. 2021, 12, 748566.
- Rosenbalm, K.E.; Tiemeyer, M.; Wells, L.; Aoki, K.; Zhao, P. Glycomics-informed glycoproteomic analysis of site-specific glycosylation for SARS-CoV-2 spike protein. STAR Protoc. 2020, 1, 100214.
- Grant, O.C.; Montgomery, D.; Ito, K.; Woods, R.J. Analysis of the SARS-CoV-2 spike protein glycan shield: Implications for immune recognition. Sci. Rep. 2020, 10, 14991.
- Zhao, P.; Praissman, J.L.; Grant, O.C.; Cai, Y.; Xiao, T.; Rosenbalm, K.E.; Aoki, K.; Kellman, B.P.; Bridger, R.; Barouch, D.H.; et al. Virus-Receptor Interactions of Glycosylated SARS-CoV-2 Spike and Human ACE2 Receptor. Cell Host Microbe 2020, 28, 586–601.e6.
- Zhao, X.; Chen, H.; Wang, H. Glycans of SARS-CoV-2 Spike Protein in Virus Infection and Antibody Production. Front. Mol. Biosci. 2021, 8, 629873.
- Wang, Y.; Wu, Z.; Hu, W.; Hao, P.; Yang, S. Impact of Expressing Cells on Glycosylation and Glycan of the SARS-CoV-2 Spike Glycoprotein. ACS Omega 2021, 6, 15988–15999.
- Kasuga, Y.; Zhu, B.; Jang, K.-J.; Yoo, J.-S. Innate immune sensing of coronavirus and viral evasion strategies. Exp. Mol. Med. 2021, 53, 723–736.
- Bieberich, E. Sphingolipids and lipid rafts: Novel concepts and methods of analysis. Chem. Phys. Lipids 2018, 216, 114–131.
- Törnquist, K.; Asghar, M.Y.; Srinivasan, V.; Korhonen, L.; Lindholm, D. Sphingolipids as Modulators of SARS-CoV-2 Infection. Front. Cell Dev. Biol. 2021, 9, 689854.
- Caterino, M.; Gelzo, M.; Sol, S.; Fedele, R.; Annunziata, A.; Calabrese, C.; Fiorentino, G.; D’Abbraccio, M.; Dell’Isola, C.; Fusco, F.M.; et al. Dysregulation of lipid metabolism and pathological inflammation in patients with COVID-19. Sci. Rep. 2021, 11, 2941.
- Meoni, G.; Ghini, V.; Maggi, L.; Vignoli, A.; Mazzoni, A.; Salvati, L.; Capone, M.; Vanni, A.; Tenori, L.; Fontanari, P.; et al. Metabolomic/lipidomic profiling of COVID-19 and individual response to tocilizumab. PLoS Pathog. 2021, 17, e1009243.
- Spick, M.; Longman, K.; Frampas, C.; Lewis, H.; Costa, C.; Walters, D.D.; Stewart, A.; Wilde, M.; Greener, D.; Evetts, G.; et al. Changes to the sebum lipidome upon COVID-19 infection observed via rapid sampling from the skin. EClinicalMedicine 2021, 33, 100786.
- Bai, Y.; Huang, W.; Li, Y.; Lai, C.; Huang, S.; Wang, G.; He, Y.; Hu, L.; Chen, C. Lipidomic alteration of plasma in cured COVID-19 patients using ultra high-performance liquid chromatography with high-resolution mass spectrometry. Biosci. Rep. 2021, 41, BSR20204305.
- Bruzzone, C.; Bizkarguenaga, M.; Gil-Redondo, R.; Diercks, T.; Arana, E.; de Vicuña, A.G.; Seco, M.; Bosch, A.; Palazón, A.; San Juan, I.; et al. SARS-CoV-2 Infection Dysregulates the Metabolomic and Lipidomic Profiles of Serum. iScience 2020, 23, 101645.
