MicroRNAs (miRNAs), short-chain RNAs of 18–22 nt chain length, are expressed in all vertebrates and control tissue development, differentiation, cell growth, and apoptosis in non-cancer and cancer cells. Additionally, miRNAs target mRNAs on the basis of their sequence and decrease protein production by inhibiting mRNA translation or destroying the mRNAs, thereby controlling cellular homeostasis. miRNA expression is controlled through DNA modification, such as by methylation and transcriptional factors through the signal-transduction pathway. miRNAs are transcribed as pri-mature from DNA and are then processed by the Drosha complex, thus generating pre-miRNAs. These pre-miRNAs are transported by exportin 5 from the nucleus to the cytoplasm and further processed by Dicer to form the double-stranded miRNA RNA-induced silencing complex (RISC). RISCs involving single-strand miRNAs, comprising Ago, Dicer, and trans-activation-responsive RNA-binding protein (TRBP) 2, are directed to the mRNA targets, thus regulating protein expressions. To date, the bioinformatics databases TargetScan (http://www.targetscan.org/vert_72/, 26 September 2021) and miRTarBase (http://mirtarbase.cuhk.edu.cn/php/index.php, 26 September 2021) have been used to predict the interaction between miRNAs and target mRNAs.
- microRNA
- RNA-binding protein
1. RNA-Binding Proteins
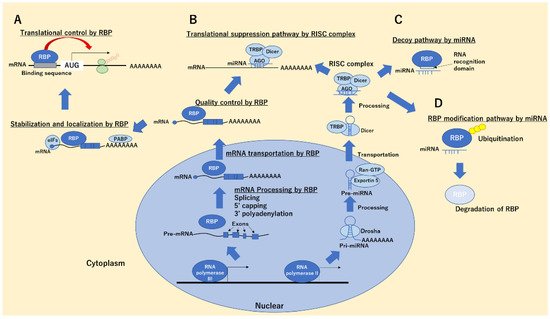
2. RBPs in Cancer Progression and Suppression
3. miRNA–RBP Binding Functions in Cancer Cells
| miRNA | Type of Pathway | Target | Type of Cancer |
Function | Reference |
|---|---|---|---|---|---|
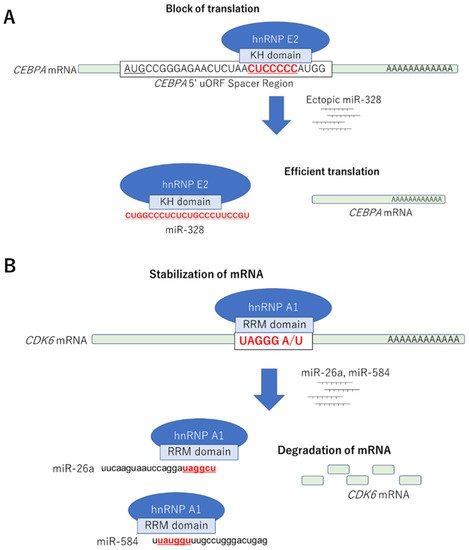
| miR-328 | Decoy | hnRNP E2-CEBPα mRNA | Leukemic blasts | Differentiation | Eiring AM, Cell 2010 |
4. Strategies for Identifying Interactions between miRNAs and RBPs
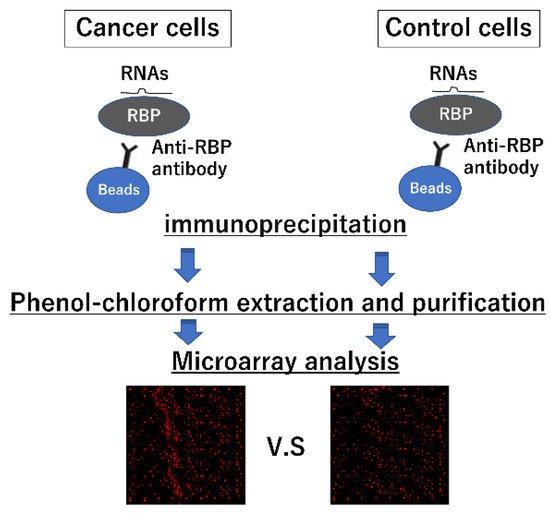
| miRs with Greater than 4-Fold Expression Compared to the Isotype Control IgG | |||||||
|---|---|---|---|---|---|---|---|
| Name | ID | Ratio (hnRNP A1/IgG) | LOG2ratio | ||||
| hsa-miR-29a-3p | MIMAT0000086 | 11.49 | 3.52 | ||||
| Canonical | PIM1 mRNA | Decreased survival | |||||
| hsa-miR-26a-5p | MIMAT0000082 | 11.37 | 3.51 | miR-29 | Decoy | ||
| hsa-miR-584-5p | MIMAT0003249 | HuR-A20 mRNA | Sarcoma | Differentiation | Balkhi MY, Sci signal, 2013 | ||
| 9.93 | 3.31 | miR-26a, -584 |
Decoy | hnRNP A1-CDK6 mRNA | Colorectal cancer | ||
| hsa-miR-107 | MIMAT0000104 | Cell growth suppression | Konishi H, Biochem Biophys Res Commun. 2015 | ||||
| 9.73 | 3.28 | miR-574-3p | Decoy | hnRNP L-VEGFA mRNA | Myeloid cells | Inhibition of cell proliferation | Yao P, Nucleic Acids Research, 2017 |
| Canonical | EP300 mRNA | ||||||
| miR-574-5p | Decoy | CUGBP1-mPGES-1 mRNA | Lung tumor | Cell growth promotion | Saul MJ, FASEB J, 2019 | ||
| miR-18a | Degradation | hnRNP A1 | Colorectal cancer | Apoptosis induction | Fujiya M, Oncogene, 2014 | ||
| hsa-miR-106b-5p | |||
| MIMAT0000680 | |||
| 8.99 | 3.17 | ||
| hsa-miR-1229-5p | MIMAT0022942 | 8.88 | 3.15 |
| hsa-miR-29b-3p | MIMAT0000100 | 8.07 | 3.01 |
| hsa-miR-194-5p | MIMAT0000460 | 8.07 | 3.01 |
| hsa-miR-142-3p | MIMAT0000434 | 7.97 | 2.99 |
| hsa-miR-18a-5p | MIMAT0000072 | 7.93 | 2.99 |
| hsa-let-7c-5p | MIMAT0000064 | 7.31 | 2.87 |
| hsa-miR-16-5p | MIMAT0000069 | 7.22 | 2.85 |
| hsa-miR-500a-3p | MIMAT0002871 | 6.96 | 2.8 |
| hsa-miR-200b-3p | MIMAT0000318 | 6.89 | 2.78 |
| hsa-miR-19a-3p | MIMAT0000073 | 6.55 | 2.71 |
| hsa-miR-222-3p | MIMAT0000279 | 6.4 | 2.68 |
| hsa-let-7b-5p | MIMAT0000063 | 5.98 | 2.58 |
| hsa-miR-23a-3p | MIMAT0000078 | 5.97 | 2.58 |
| hsa-let-7d-5p | MIMAT0000065 | 5.85 | 2.55 |
| hsa-miR-431-3p | MIMAT0004757 | 5.7 | 2.51 |
| hsa-miR-200c-3p | MIMAT0000617 | 5.62 | 2.49 |
| hsa-miR-23b-3p | MIMAT0000418 | 5.27 | 2.4 |
| hsa-miR-27b-3p | MIMAT0000419 | 5.23 | 2.39 |
| hsa-miR-19b-3p | MIMAT0000074 | 5.05 | 2.34 |
| hsa-miR-103a-3p | MIMAT0000101 | 4.95 | 2.31 |
| hsa-miR-1246 | MIMAT0005898 | 4.73 | 2.24 |
| hsa-let-7a-5p | MIMAT0000062 | 4.66 | 2.22 |
| hsa-miR-20a-5p | MIMAT0000075 | 4.5 | 2.17 |
| hsa-miR-27a-3p | MIMAT0000084 | 4.49 | 2.17 |
| hsa-miR-141-3p | MIMAT0000432 | 4.32 | 2.11 |
| hsa-miR-21-5p | MIMAT0000076 | 4.25 | 2.09 |
| hsa-miR-17-5p | MIMAT0000070 | 4.24 | 2.08 |
| hsa-miR-106a-5p | MIMAT0000103 | 4.15 | 2.05 |
| hsa-miR-20b-5p | MIMAT0001413 | 4.11 | 2.04 |
