Serotonin (Ser) and melatonin (Mel) serve as master regulators of plant growth and development by influencing diverse cellular processes. The enzymes namely, tryptophan decarboxylase (TDC) and tryptamine 5-hydroxylase (T5H) catalyse the formation of Ser from tryptophan. Subsequently, serotonin N-acetyl transferase (SNAT) and acetyl-serotonin methyltransferase (ASMT) form Mel from Ser. Plant genomes harbour multiple genes for each of these four enzymes, all of which have not been identified yet.
- abiotic stress
- Arabidopsis
- melatonin
- miRNA
- rice
- serotonin
- sorghum
- tomato
[1][2][3][4][5][6][7][8][9][10][11][12][13][14][15][16][17][18][19][20][21][22][23][24][25][26][27][28][29][30][31][32][33][34][35][36][37][38][39][40][41][42][43][44][45][46][47][48][49][50][51][52][53][54][55][56][57][58][59][60][61][62][63][64][65][66][67][68][69][70][71][72][73][74][75]
1. Introduction
Serotonin (Ser) and melatonin (Mel) are indoleamines which are synthesised in most life forms including prokaryotes such as bacteria, as well as fungi, nematodes, animals and plants; however, their presence in archaea remains to be studied [1]. They are well known neurotransmitters, which play vital roles, especially in animals, being associated with diverse physiological processes. In humans, Ser governs behavioural patterns, i.e., moods and appetite, and Mel regulates sleep, circadian rhythms, immunity enhancement and even aids in the reduction of oxidative stress in animals [2,3][2][3]. With the emergence of plant neurobiology as an exciting field of research, investigations pertaining to the precise roles of Ser and Mel in plants have received greater attention [4].
Tryptophan, an essential aromatic amino acid, primarily acts as the common substrate for biosynthesis of various plant indoleamines, namely, Ser, Mel and auxin [5,6,7][5][6][7]. Cells containing high tryptophan levels, generally, accumulate greater amounts of Ser and, subsequently, Mel [8]. According to the classical pathway, tryptophan decarboxylase (TDC) and tryptamine 5-hydroxylase (T5H) enzymes are required for the production of Ser, whereas serotonin N-acetyltransferase (SNAT) and acetyl serotonin methyltransferase (ASMT) enzymes are required for Mel production from Ser in plants (Figure 1). While TDC is the rate-limiting enzyme of Ser biosynthesis, both SNAT and ASMT SNAT and ASMT regulate Mel synthesis under specific conditions and catalyse the dedicated steps in Mel biosynthesis [3,8][3][8]. The SNAT enzyme is regulated by feedback inhibition in rice [9] (Figure 1). In silico studies pertaining to the genes involved in Mel biosynthesis have led to the identification of 5 genes encoding for TDC proteins and 14 genes encoding for ASMT ASMT proteins in the tomato genome [10,11][10][11]. Furthermore, the existence of TDC in in Citrus is also experimentally proven [12].
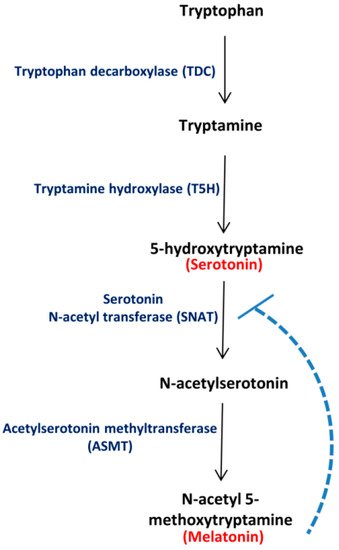
Figure 1. Major route for Ser and Mel biosynthesis in plants. Tryptophan decarboxylase (TDC) converts tryptophan to tryptamine, which is converted to 5-hydroxytryptamine (Ser) by tryptamine 5-hydroxylase (T5H). Serotonin N-acetyl transferase (SNAT) converts Ser to N-acetyl serotonin, which is further transformed to N-acetyl 5-methoxytryptamine (Mel) by acetylserotonin methyltransferase (ASMT). The broken line depicts feedback regulation of SNAT by Mel, as reported in literature.
Notably, light/dark conditions can modulate Mel levels in plants. In herbaceous peony, Mel production depends on light availability, specifically increasing at the bloom stage of its development [13]. This increase, in turn, can be correlated with enhanced gene expression patterns of the rate-limiting enzyme, TDC [13] [13]. Likewise, expression levels of genes encoding for Ser and Mel biosynthetic enzymes in rice are also altered during light and dark conditions [14,15][14][15].
Mel regulates multiple plant physiological pathways such as growth and reproduction, senescence, chrono-regulation, root and shoot organogenesis and diverse hormonal crosstalk, as well as stress responses [16,17,18,19][16][17][18][19]. It acts as a scavenger of reactive oxygen species, reduces cellular damage in plants by repairing mitochondria and improves the shelf life of fruits and vegetables, owing to its antioxidant properties [20,21,22,23][20][21][22][23]. The multifunctional roles of Mel in plants and the recent discovery of the first plant Mel receptor has led to emerging speculations of Mel acting as a new phytohormone or master regulator [24]. Likewise, Ser also plays diverse roles in plants, with its functions in stress response being delineated in the last decade [7,25,26][7][25][26].
Importantly, the role of Ser and Mel in mediating stress-related signalling is also emerging [19,27,28,29,30][19][27][28][29][30]. Ser and Mel have been shown to regulate root growth in sunflower under salinity stress [31] [31]. Interestingly, nitric oxide in conjunction with Mel modulates glutathione reductase activity and alters glutathione content in sunflower seedling cotyledons under salinity stress conditions [32]. Indeed, Mel along with nitric oxide acts as a salt-signalling molecule in sunflower and mediates stress responses via its antioxidant property [33]. In addition, microRNAs (miRNAs) have also been found to modulate abiotic stress responses in plants [34]. Furthermore, Mel in conjunction with a specific miRNA, miR398, has been found to mediate oxidative and heavy metal stress tolerance in plants [35].
There are several reports highlighting the roles of Ser and Mel in plant development and stress responses. However, no systematic information regarding the genetic organization of the respective genes involved in Ser and Mel biosynthesis is presently available in plants. Hence, in the current study, we have attempted to identify the genes involved in Ser and Mel biosynthesis in four model plant species, namely, Arabidopsis, rice, sorghum and tomato, followed by analysis of their evolutionary relationships. Gene expression profiles of genes encoding enzymes for Ser and Mel biosynthesis, in different tissues/organs during various stages of development, under light/dark conditions and in response to various stress factors, were also analysed. Additionally, we could even identify miRNAs that regulate the expression of genes encoding enzymes for Ser and Mel biosynthesis in selected model plants. Taken together, we provide detailed information regarding gene family members associated with Ser and Mel biosynthesis in plants. Our study can be beneficial for identifying the crucial targets needed for manipulation of Ser and Mel pathway.
2. Distribution and regulation of genes encoding Ser and Mel biosynthetic enzymes in plants
In order to compensate for the lack of mobility, plants have evolved an intricate network of signals which sense and respond to the surrounding environment. Intriguingly, despite being devoid of a central nervous system, plants are still capable of producing certain signalling molecules similar to those found in the mammalian nervous system, referred to as neurotransmitters. Ser (or 5-hydroxy-tryptamine) and Mel (or N-Acetyl-5-methoxytryptamine) are examples of the same. These have been recognized as novel plant growth regulators possessing a plethora of physiological functions in plants, ranging from development to abiotic stress responses [28,36][28][36]. These molecules are in direct crosstalk with several plant hormones and interrelated with crucial signalling pathways [18,36][18][36]. Considering the important, diverse and multifaceted roles of these signalling molecules in plant life, it is worth identifying genes encoding for enzymes mediating Ser and Mel biosynthetic pathway in plants. The major biosynthetic pathway for these indoleamines includes four enzymes, namely, tryptophan decarboxylase (TDC), tryptamine 5-hydroxylase (T5H), serotonin N-acetyl transferase (SNAT) and acetyl-serotonin methyltransferase (ASMT). Of them, only selected gene families have been functionally characterised to date, such as TDCs in tomato, citrus and rice, and ASMTs in tomato and pepper [8,10,11,12,37][8][10][11][12][37]. Our comprehensive genome-wide analysis led to the identification of possible gene family members of all the four enzymes of the Ser and Mel biosynthetic pathway in rice, Arabidopsis, tomato and sorghum (Figure 2). We found that 7 and 9 TDC genes, 2 and 1 SNAT genes and 19 and 28 ASMT genes are present in rice and sorghum, respectively In Arabidopsis and tomato, 2 and 5 TDC genes, 1 and 2 SNAT genes and 17 and 11 ASMT genes were respectively identified. On the other hand, T5H is present as a single gene in rice, sorghum and Arabidopsis, whereas in tomato, six isoforms of the T5H gene could be detected. Our phylogenetic analysis revealed each enzyme to be clustered into different groups, consisting of proteins from either the same or different species, indicating diversification of the proteins.
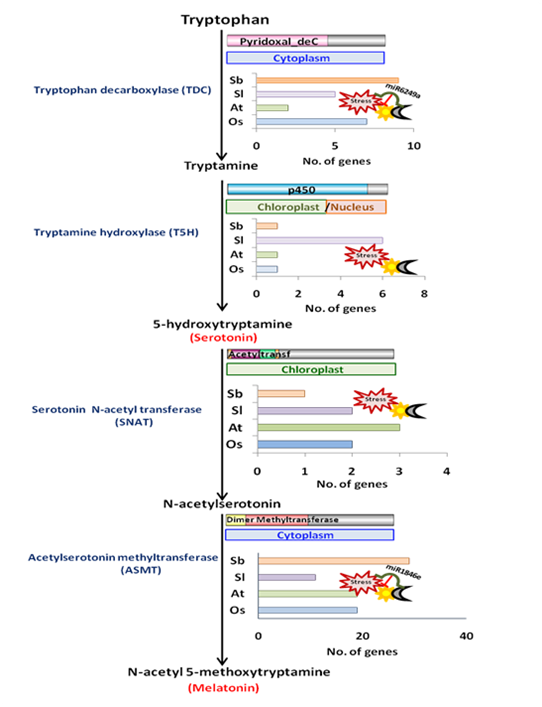
Figure 2. Diagrammatic representation of the distribution and regulation of genes encoding Ser and Mel biosynthetic enzymes in plants.
The presence of Ser and Mel is ubiquitous [3] and is also conserved across plants [38]. Chloroplasts and mitochondria are considered to be the original sites for the synthesis of Ser and Mel in plants, inheriting Ser and Mel biosynthetic abilities from their bacterial ancestor [39]. In particular, the presence of genes encoding Ser and Mel biosynthetic enzymes in alpha-proteobacteria and cyanobacteria is important, as they are considered to be the ancestors of the plant organelles. In correlation with this fact, SNAT proteins from rice, one of the key enzymes in Mel biosynthesis, shows substantial homology at the gene level to that of the cyanobacterium Synechocystis and, likewise, is plastidially located [40]. In fact, all SNAT proteins characterized to date are strictly chloroplastic, reaffirming their cyanobacterial heritage. However, our analysis has suggested that different genes of the Ser and Mel biosynthetic pathway may have variable localization. While TDC TDC genes majorly localise in the cytoplasm (except for Arabidopsis TDCs, which were mostly chloroplastic), both T5H and SNAT T5H and SNAT genes were predicted in the chloroplast in all four plant species, in agreement with the endosymbiosis theory. ASMTs, however, exhibited highly variable sub-cellular localization patterns within the four plant species. While members of the AtASMT and and SbASMT gene family were predicted to be cytoplasmic, only a very few members of the OsASMT and and SlASMT gene family showed similar localization patterns. Some members of the rice and tomato ASMT ASMT gene family were indeed found to localize in the cytoskeleton and nucleus. Preliminary work in rice and other recent reports also show localization of the biosynthetic enzymes in cytoplasm, chloroplast and mitochondria [9,40,41][9][40][41].
Currently, limited information is available regarding the tissue-specificity of Ser. However, there is some evidence to suggest that its levels are highest in the aerial parts of plants, such as in potato and Ligisticumspp [42,43] [42][43]. In addition, Ramakrishna et al. [44] found Ser to be localised in the mature roots and stems of Coffea canephora Pierre ex. Using quantum dots, Mel and Ser uptake in the axenic roots of St. John’s wort has also been studied. Under optimal conditions, Mel is absorbed through epidermal cells, travels laterally across the root cortex and accumulates in endodermal tissues and rapidly dividing pericycle cells, while Ser is absorbed by cells proximal to the crown and is rapidly transported towards the root tip [45]. Our analysis of the transcript abundance of putative TDC and T5H and T5H genes involved in Ser biosynthesis reveals lesser tissue-specific variations across the four different plant species. In fact, OsT5H1 and OsTDC5 from rice are constitutively expressed in all the tissues. Likewise, the SNAT and ASMT genes involved in Mel biosynthesis are also expressed in all tissue types in rice, Arabidopsis, sorghum and tomato. While the expression levels of some genes vary across tissues, others , showed constitutive expression, suggesting Mel to be distributed throughout the plant organs, an observation which is supported by studies on various edible and medicinal plants [46,47,48][46][47][48].
Ser and Mel probably exist in a balance, similar to that established for auxin and cytokinin, due to their close biosynthetic relationship. While Mel is suggested to behave in a similar manner to auxin, Ser acts in the same way as cytokinin, implicating its role in organogenesis, morphogenesis and the growth and development of plants [6]. Furthermore, transcript levels of the majority of genes encoding Ser and Mel biosynthetic enzymes are altered at specific developmental stages. For instance, AtTDC1 from Arabidopsis is upregulated at the seed germination stage, SlT5H6 from tomato at the fruit ripening stage and OsTDC1 from rice at the heading stage, whereas SbASMT14 is specifically induced at the booting stage of development. However, some genes, such as AtASMT7 and OsT5H1, are induced at all stages of plant development. Besides flowering and morphogenesis, the role of Ser is also well established in senescence [49]. In fact, Kang et al. [50] reported enhanced accumulation of Ser in rice leaves undergoing senescence, its synthesis being closely coupled with the transcriptional induction of its biosynthetic genes, especially TDC. In agreement with this, we found 2 AtTDC genes to be induced under senescence. Notably, ASMT genesalso showed enhanced expression under senescence. This is particularly interesting because only very few studies have shown the regulation of Mel biosynthesis in promoting senescence [14,51][14][51]. Most studies have focussed on the exogenous application of Mel in delaying aging [52,53,54][52][53][54]. The senescence-specific induction of genes encoding Ser and Mel biosynthetic enzymes, therefore, suggests the specific role of Ser and Mel in senescence.
Furthermore, Ser biosynthesis is differentially induced in response to different wavelengths of light [55]. Exposure to red light leads to immediate accumulation of Ser in Scutellaria, which decreases over 7 days. In contrast, Mel is detected in plants grown under blue light [56]. Using a short-day flowering plant, Chenopodium rubrum, Kolářet al. [57] showed that light suppresses Mel biosynthesis in plants. However, Murch et al. [58] reported that light significantly stimulates Mel production in St. John’s wort. Our assessment of rice Ser and Mel encoding genes through qRT-PCR are in agreement with the results of Murch et al. [58]. In our analysis, we found all members of the OsTDC, OsT5H, OsSNAT and and OsASMT gene families to be up-regulated in response to 6h light exposure subsequent to prolonged (for 2 d) darkness. On the contrary, data retrieved from public repositories indicate OsSNAT and and OsASMT expression to be more pronounced in darkness. We believe that a change of growth cycle in our experiment has led to this difference in regulation of Ser and Mel biosynthetic genes as short/long day light cycles also affect Mel accumulation. In fact, the endogenous levels of Mel have been shown to vary in a typical diurnal pattern in Arabidopsis [28].
Furthermore, Ser and Mel also play an important role in imparting stress tolerance to plants (Figure 3). Ser and Mel mediate abiotic stress tolerance via their antioxidative defensive role, which leads to the detoxification of ROS and reactive nitrogen species (RNS) [59,60,61][59][60][61]. There is a wealth of information related to the effects of exogenous application of Ser and Mel in mitigating abiotic stresses in plants [32,62,63,64,65][32][62][63][64][65]. We also found genes encoding Ser and Mel biosynthetic enzymes to be maximally induced in response to drought conditions as well as upon ABA treatment, thereby reinforcing the role of ABA in the regulation of Ser and Mel levels under drought conditions. In addition, the available literature also highlights the role of these metabolites in biotic stresses such as herbivory and pathogen attack [66,67,68][66][67][68]. Mel induces SA and NO biosynthesis, acting upstream of the defence signalling pathway, thereby stimulating disease resistance because of co-action [69]. In agreement with this, we found most of the genes encoding enzymes for Ser and Mel biosynthesis in rice to be upregulated in response to both biotic stress and JA.

Figure 3. Model summarizing the members of Ser and Mel biosynthetic pathway of rice, highlighting their regulation. The crosstalk between genes encoding enzymes for Ser and Mel biosynthesis (identified by genome-wide analysis) harboured in different sub-cellular compartments, have been illustrated under stress conditions, based on transcript profile validations. The miRNA-based regulation of specific gene family members has also been indicated. The black dotted arrows depict the stress-inducibility of specific transcripts regulating Ser and Mel biosynthesis under various stress conditions, as validated by qRT-PCR. Solid black lines depict regulation of transcripts with specific miRNAs. Coloured arrows depict the genes encoding the enzymes involved in the Ser and Mel biosynthesis.
Importantly, the response of Ser and Mel biosynthetic genes to hormone treatments was found to be highly variable across four species. While ABA induced gene expression in monocots, the same had a repressive effect in Arabidopsis. However, GA showed a reverse response in both species, where it repressed gene expression in rice but induced it in Arabidopsis. Previous studies report that Mel alters both these hormone levels [18]. It up-regulates ABA catabolism in cucumber seedlings (under salinity) and apple leaves (under drought), whereas it increases ABA content in the monocot barley (under drought/cold stress) [70,71,72][70][71][72]. On the other hand, GA biosynthesis is enhanced in cucumber seedlings in response to Mel [70,71][70][71]. Thus, an intricate feedback regulatory mechanism exists in plants to control Ser and Mel levels.
Interestingly, miRNAs are also involved in the regulation of Ser and Mel gene expression in plants. Mel is known to regulate miRNA gene expression in plants under certain conditions [73], but no report of miRNA–mediated regulation of Ser and Mel gene expression is yet available. Nevertheless, we could predict 51 unique miRNAs regulating Ser and Mel biosynthetic genes across the four plant species, of which, miR156 was found to be conserved in all species except Arabidopsis. However, some miRNAs, such as miR393 and miR398, could be identified in only Arabidopsis, suggesting that, possibly, miRNA regulation of these genes either originated relatively recently or was gradually lost in some of the plant species. The miR398 is also known to mediate oxidative and heavy-metal stress tolerance in plants [34[34][35],35], whereas miR393 has been shown to target the ABF2/TIR transcription factor, which in turn regulates auxin biosynthesis [74]. Furthermore, we found OsTDC5 and and OsASMT18 from rice to be regulated by miR6249a and miR1846e, respectively, under light/dark conditions and even under salinity and drought stress. Interestingly, both these miRNAs have been reported to be drought-responsive [75]; miROs1846e was indeed found to positively regulate OsASMT18 under drought conditions.
Therefore, through this study, we have been able to gain crucial insights into the Ser and Mel biosynthesis pathway in plants. Indeed, expression patterns of these biosynthetic genes is regulated by stress conditions in plants and even exhibits tissue and developmental variations, in corroboration with diversity in their actions. Furthermore, similar to animals, these genes also exhibit light/dark cycle-mediated alterations in plants. Collectively, Figure 3 summarizes the information regarding rice genes encoding Ser and Mel biosynthetic enzymes and their regulation. Our study has, thus, generated a knowledge repository and paved the way for future investigations, which will enable us to gain deeper insights into the diverse regulatory roles of genes encoding enzymes for Ser and Mel biosynthesis in selected model plants.
References
- Tan, D.X.; Hardeland, R.; Back, K.; Manchester, L.C.; Alatorre-Jimenez, M.A.; Reiter, R.J. On the significance of an alternate pathway of melatonin synthesis via 5-methoxytryptomine: Comparisons across species. J. Pineal Res. 2016, 61, 27–40. [Google Scholar] [CrossRef]
- Lv, J.; Liu, F. The role of serotonin beyond the central nervous system during embryogenesis. Front. Cell. Neurosci. 2017, 11, 74. [Google Scholar] [CrossRef]
- Zhao, D.; Yu, Y.; Shen, Y.; Liu, Q.; Zhao, Z.; Sharma, R.; Reiter, R.J. Melatonin Synthesis and Function: Evolutionary History in Animals and Plants. Front. Endocrinol. 2019, 10, 249. [Google Scholar] [CrossRef]
- Iriti, M. Plant Neurobiology, a fascinating perspective in the field of research on plant secondary metabolites. Int. J. Mol. Sci. 2013, 14, 10819–10821. [Google Scholar] [CrossRef]
- Pelagio-Flores, R.; Lopez-Bucio, J. Serotonin and melatonin in plant growth and development. In Serotonin and Melatonin: Their Functional Role in Plants, Food, Phytomedicine, and Human Health; Ravishankar, G.A., Ramakrishna, A., Eds.; CRC Press: Boca Raton, FL, USA, 2016; pp. 97–110. [Google Scholar]
- Erland, L.A.E.; Saxena, P.K. Mammalian neurotransmitters are important signals mediating plant morphogenesis. In Sensory Biology of Plants; Sopory, S., Ed.; Springer Nature: Singapore, 2019; pp. 411–449. [Google Scholar]
- Erland, L.A.E.; Turi, C.E.; Saxena, P.K. Serotonin in Plants: Origin, Functions and Implications. In Serotonin; Pilowsky, P.M., Ed.; Elsevier Inc.: Amsterdam, The Netherlands, 2019; pp. 23–46. [Google Scholar]
- Kang, K.; Kang, S.; Lee, K.; Park, M.; Back, K. Enzymatic features of serotonin biosynthetic enzymes and serotonin biosynthesis in plants. Plant Signal. Behav. 2008, 3, 389–390. [Google Scholar] [CrossRef] [PubMed]
- Back, K.; Tan, D.X.; Reiter, R.J. Melatonin biosynthesis in plants: Multiple pathways catalyze tryptophan to melatonin in the cytoplasm or chloroplasts. J. Pineal Res. 2016, 61, 426–437. [Google Scholar] [CrossRef]
- Liu, W.; Zhao, D.; Zheng, C.; Chen, C.; Peng, X.; Cheng, Y.; Wan, H. Genomic analysis of the ASMT gene family in Solanumlycopersicum. Molecules 2017, 22, 1984. [Google Scholar] [CrossRef] [PubMed]
- Pang, X.; Wei, Y.; Cheng, Y.; Pan, L.; Ye, Q.; Wang, R.; Ruan, M.; Zhou, G.; Yao, Z.; Li, Z.; et al. The tryptophan decarboxylase in Solanumlycopersicum. Molecules 2018, 23, 998. [Google Scholar] [CrossRef] [PubMed]
- De Masi, L.; Castaldo, D.; Pignone, D.; Servillo, L.; Facchiano, A. Experimental evidence and in silico identification of tryptophan decarboxylase in citrus genus. Molecules 2017, 22, 272. [Google Scholar] [CrossRef]
- Zhao, D.; Wang, R.; Liu, D.; Wu, Y.; Sun, J.; Tao, J. Melatonin and expression of tryptophan decarboxylase gene (TDC) in Herbaceous Peony (Paeonialactiflora Pall.) flowers. Molecules 2018, 23, 1164. [Google Scholar] [CrossRef] [PubMed]
- Byeon, Y.; Park, S.; Kim, Y.-S.; Park, D.-H.; Lee, S.; Back, K. Light-regulated melatonin biosynthesis in rice during the senescence process in detached leaves. J. Pineal Res. 2012, 53, 107–111. [Google Scholar] [CrossRef]
- Wei, Y.; Zeng, H.; Hu, W.; Chen, L.; He, C.; Shi, H. Comparative transcriptional profiling of melatonin synthesis and catabolic genes indicates the possible role of melatonin in developmental and stress responses in rice. Front. Plant Sci. 2016, 7, 676. [Google Scholar] [CrossRef] [PubMed]
- Reiter, R.J.; Tan, D.-X.; Zhou, Z.; Cruz, M.H.C.; Fuentes-Broto, L.; Galano, A. Phytomelatonin: Assisting Plants to Survive and Thrive. Molecules 2015, 20, 7396–7437. [Google Scholar] [CrossRef] [PubMed]
- Erland, L.A.E.; Saxena, P.K. Melatonin in plant morphogenesis. In Vitro Cell. Dev. Biol. Plant. 2017, 54, 3–24. [Google Scholar] [CrossRef]
- Arnao, M.B.; Ruiz, J.H. Melatonin and its relationship to plant hormones. Ann. Bot. 2018, 121, 195–207. [Google Scholar] [CrossRef]
- Mukherjee, S. Novel perspectives on the molecular crosstalk mechanisms of serotonin and melatonin in plants. Plant Physiol. Biochem. 2018, 132, 33–45. [Google Scholar] [CrossRef] [PubMed]
- Fan, J.; Xie, Y.; Zhang, Z.; Chen, L. Melatonin: A Multifunctional Factor in Plants. Int. J. Mol. Sci. 2018, 19, 1528. [Google Scholar] [CrossRef] [PubMed]
- Sharif, R.; Xie, C.; Zhang, H.; Arnao, M.B.; Ali, M.; Ali, Q.; Muhammad, I.; Shalmani, A.; Nawaz, M.A.; Chen, P.; et al. Melatonin and Its Effects on Plant Systems. Molecules 2018, 23, 2352. [Google Scholar] [CrossRef]
- Li, Z.-G.; Xu, Y.; Bai, L.-K.; Zhang, S.-Y.; Wang, Y. Melatonin enhances thermotolerance of maize seedlings (Zea mays L.) by modulating antioxidant defense, methylglyoxal detoxification, and osmoregulation systems. Protoplasma 2018, 256, 471–490. [Google Scholar] [CrossRef]
- Arnao, M.B.; Hernández-Ruiz, J. The Potential of Phytomelatonin as a Nutraceutical. Molecules 2018, 23, 238. [Google Scholar] [CrossRef]
- Arnao, M.B.; Ruiz, J.H. Melatonin: A New Plant Hormone and/or a Plant Master Regulator? Trends Plant Sci. 2019, 24, 38–48. [Google Scholar] [CrossRef]
- Ramakrishna, A.; Giridhar, P.; Ravishankar, G.A. Phytoserotonin. Plant Signal. Behav. 2011, 6, 800–809. [Google Scholar]
- Erland, L.A.E.; Saxena, P.K. Beyond a neurotransmitter: The role of serotonin in plants. Neurotransmitter 2017, 4, 1538. [Google Scholar] [CrossRef]
- Kaur, H.; Mukherjee, S.; Baluska, F.; Bhatla, S.C. Regulatory roles of serotonin and melatonin in abiotic stress tolerance in plants. Plant Signal. Behav. 2015, 10, e1049788. [Google Scholar] [CrossRef] [PubMed]
- Shi, H.; Wei, Y.; He, C. Melatonin-induced CBF/DREB1s are essential for diurnal change of disease resistance and CCA1 expression in Arabidopsis. Plant Physiol. Biochem. 2016, 100, 150–155. [Google Scholar] [CrossRef] [PubMed]
- Mandal, M.K.; Suren, H.; Ward, B.; Boroujerdi, A.; Kousik, C. Differential roles of melatonin in plant-host resistance and pathogen suppression in cucurbits. J. Pineal Res. 2018, 65, e12505. [Google Scholar] [CrossRef] [PubMed]
- Bose, S.K.; Howlader, P. Melatonin plays multifunctional role in horticultural crops against environmental stresses: A review. Environ. Exp. Bot. 2020, 176, 104063. [Google Scholar] [CrossRef]
- Mukherjee, S.; David, A.; Yadav, S.; Baluška, F.; Bhatla, S.C. Salt stress-induced seedling growth inhibition coincides with differential distribution of serotonin and melatonin in sunflower seedling roots and cotyledons. Physiol. Plant. 2014, 152, 714–728. [Google Scholar] [CrossRef]
- Kaur, H.; Bhatla, S.C. Melatonin and nitric oxide modulate glutathione content and glutathione reductase activity in sunflower seedling cotyledons accompanying salt stress. Nitric Oxide 2016, 59, 42–53. [Google Scholar] [CrossRef]
- Arora, D.; Bhatla, S.C. Melatonin and nitric oxide regulate sunflower seedling growth under salt stress accompanying differential expression of Cu/Zn SOD and Mn SOD. Free. Radic. Biol. Med. 2017, 106, 315–328. [Google Scholar] [CrossRef]
- Tripathi, A.; Goswami, K.; Sanan-Mishra, N. Role of bioinformatics in establishing microRNAs as modulators of abiotic stress responses: The new revolution. Front. Physiol. 2015, 6, 286. [Google Scholar] [CrossRef] [PubMed]
- Gu, Q.; Chen, Z.; Yu, X.; Cui, W.; Pan, J.; Zhao, G.; Xu, S.; Wang, R.; Shen, W. Melatonin confers plant tolerance against cadmium stress via the decrease of cadmium accumulation and reestablishment of microRNA-mediated redox homeostasis. Plant Sci. 2017, 261, 28–37. [Google Scholar] [CrossRef]
- Erland, L.; Murch, S.J.; Reiter, R.J.; Saxena, P.K. A new balancing act: The many roles of melatonin and serotonin in plant growth and development. Plant Signal. Behav. 2015, 10, e1096469. [Google Scholar] [CrossRef] [PubMed]
- Pan, L.; Zheng, J.; Liu, J.; Guo, J.; Liu, F.; Liu, L.H. Analysis of the ASMT gene family in pepper (Capsicum annuum L.): Identification, phylogeny, and expression profiles. Int. J. Genom. 2019, 2019, 7241096. [Google Scholar] [CrossRef] [PubMed]
- Tan, D.-X.; Hardeland, R.; Manchester, L.C.; Korkmaz, A.; Ma, S.; Rosales-Corral, S.; Reiter, R.J. Functional roles of melatonin in plants, and perspectives in nutritional and agricultural science. J. Exp. Bot. 2012, 63, 577–597. [Google Scholar] [CrossRef]
- Tan, D.-X.; Reiter, R.J. An evolutionary view of melatonin synthesis and metabolism related to its biological functions in plants. J. Exp. Bot. 2020, 71, 4677–4689. [Google Scholar] [CrossRef]
- Byeon, Y.; Lee, K.; Park, Y.I.; Park, S.; Back, K. Molecular cloning and functional analysis of serotonin N-acetyltransferase from the cyanobacterium Synechocystis sp. PCC 6803. J. Pineal Res. 2013, 55, 371–376. [Google Scholar] [CrossRef]
- Byeon, Y.; Lee, H.Y.; Lee, K.; Park, S.; Back, K. Cellular localization and kinetics of the rice melatonin biosynthetic enzymes SNAT and ASMT. J. Pineal Res. 2014, 56, 107–114. [Google Scholar] [CrossRef] [PubMed]
- Engström, K.; Lundgren, L.; Samuelsson, G. Bioassay-guided isolation of serotonin from fruits of Solanum tuberosum L. Acta Pharm. Nord. 1992, 4, 91–92. [Google Scholar]
- Turi, C.E.; Murch, S.J. Targeted and Untargeted Phytochemistry of Ligusticum canbyi: Indoleamines, Phthalides, Antioxidant Potential, and Use of Metabolomics as a Hypothesis-Generating Technique for Compound Discovery. Planta Med. 2013, 79, 1370–1379. [Google Scholar] [CrossRef] [PubMed]
- Ramakrishna, A.; Giridhar, P.; Jobin, M.; Paulose, C.S.; Ravishankar, G.A. Indoleamines and calcium enhance somatic embryogenesis in Coffeacanephora P ex Fr. Plant Cell Tissue Organ Cult. 2012, 108, 267–278. [Google Scholar] [CrossRef]
- Erland, L.A.E.; Yasunaga, A.; Li, I.T.S.; Murch, S.J.; Saxena, P.K. Direct visualization of location and uptake of applied melatonin and serotonin in living tissues and their redistribution in plants in response to thermal stress. J. Pineal Res. 2018, 66, e12527. [Google Scholar] [CrossRef] [PubMed]
- Dubbels, R.; Reiter, R.J.; Klenke, E.; Goebel, A.; Schnakenberg, E.; Ehlers, C.; Schiwara, H.W.; Schloot, W. Melatonin in edible plants identified by radioimmunoassay and by high performance liquid chromatography-mass spectrometry. J. Pineal Res. 1995, 18, 28–31. [Google Scholar] [CrossRef] [PubMed]
- Badria, F.A. Melatonin, Serotonin, and Tryptamine in Some Egyptian Food and Medicinal Plants. J. Med. Food 2002, 5, 153–157. [Google Scholar] [CrossRef]
- Hernández-Ruiz, J.; Cano, A.; Arnao, M.B. Melatonin: A growth-stimulating compound present in lupin tissues. Planta 2004, 220, 140–144. [Google Scholar] [CrossRef]
- Lee, K.; Back, K. Overexpression of rice serotonin N -acetyltransferase1 in transgenic rice plants confers resistance to cadmium and senescence and increases grain yield. J. Pineal Res. 2017, 62, e12392. [Google Scholar] [CrossRef]
- Kang, K.; Kim, Y.-S.; Park, S.; Back, K. Senescence-Induced Serotonin Biosynthesis and Its Role in Delaying Senescence in Rice Leaves. Plant Physiol. 2009, 150, 1380–1393. [Google Scholar] [CrossRef] [PubMed]
- Park, S.; Lee, K.; Kim, Y.-S.; Back, K. Tryptamine 5-hydroxylase-deficient Sekiguchi rice induces synthesis of 5-hydroxytryptophan and N-acetyltryptamine but decreases melatonin biosynthesis during senescence process of detached leaves. J. Pineal Res. 2011, 52, 211–216. [Google Scholar] [CrossRef] [PubMed]
- Wang, P.; Sun, X.; Chang, C.; Feng, F.; Liang, D.; Cheng, L.; Ma, F. Delay in leaf senescence of Malushupehensis by long-term melatonin application is associated with its regulation of metabolic status and protein degradation. J. Pineal Res. 2013, 55, 424–434. [Google Scholar] [CrossRef] [PubMed]
- Liang, C.; Zheng, G.; Li, W.; Wang, Y.; Hu, B.; Wang, H.; Wu, H.; Qian, Y.; Zhu, X.-G.; Tan, D.-X.; et al. Melatonin delays leaf senescence and enhances salt stress tolerance in rice. J. Pineal Res. 2015, 59, 91–101. [Google Scholar] [CrossRef] [PubMed]
- Liang, D.; Shen, Y.; Ni, Z.; Wang, Q.; Lei, Z.; Xu, N.; Deng, Q.; Lin, L.; Wang, J.; Lv, X.; et al. Exogenous melatonin application delays senescence of kiwifruit leaves by regulating the antioxidant capacity and biosynthesis of flavonoids. Front. Plant Sci. 2018, 9, 426. [Google Scholar] [CrossRef]
- Reynolds, J.D.; Kimbrough, T.D.; Weekley, L.B. The effect of light quality on 5-hydroxyindole metabolism in leaves of Sedum morganianum (Crassulaceae). Biochem. Physiol. Pflanz. 1985, 180, 345–351. [Google Scholar] [CrossRef]
- Forsyth, J.; Erland, L.; Shipley, P.; Murch, S. Plant perception of light: The role of indoleamines in Scutellaria species. Melatonin Res. 2020, 3, 161–176. [Google Scholar] [CrossRef]
- Kolář, J.; Macháčková, I.; Eder, J.; Prinsen, E.; van Dongen, W.; van Onckelen, H.; Illnerová, H. Melatonin: Occurrence and daily rhythm in Chenopodium rubrum. Phytochemistry 1997, 44, 1407–1413. [Google Scholar] [CrossRef]
- Murch, S.; Krishnaraj, S.; Saxena, P. Tryptophan is a precursor for melatonin and serotonin biosynthesis in in-vitro regenerated St. John’s wort (Hypericumperofatum L. cv. Anthos) plants. Plant Cell Rep. 2000, 19, 698–704. [Google Scholar] [CrossRef]
- Hardeland, R. Antioxidative Protection by Melatonin: Multiplicity of Mechanisms from Radical Detoxification to Radical Avoidance. Endocrine 2005, 27, 119–130. [Google Scholar] [CrossRef]
- Yu, Y.; Lv, Y.; Shi, Y.; Li, T.; Chen, Y.; Zhao, D.; Zhao, Z. The role of phyto-melatonin and related metabolites in response to stress. Molecules 2018, 23, 1887. [Google Scholar] [CrossRef]
- Pardo-Hernández, M.; López-Delacalle, M.; Rivero, R. ROS and NO Regulation by Melatonin Under Abiotic Stress in Plants. Antioxidants 2020, 9, 1078. [Google Scholar] [CrossRef]
- Zhang, N.; Sun, Q.; Zhang, H.; Cao, Y.; Weeda, S.; Ren, S.; Guo, Y.-D. Roles of melatonin in abiotic stress resistance in plants. J. Exp. Bot. 2015, 66, 647–656. [Google Scholar] [CrossRef]
- Wang, Y.; Reiter, R.J.; Chan, Z. Phytomelatonin: A universal abiotic stress regulator. J. Exp. Bot. 2017, 69, 963–974. [Google Scholar] [CrossRef]
- Ke, Q.; Ye, J.; Wang, B.; Ren, J.; Yin, L.; Deng, X.; Wang, S. Melatonin Mitigates Salt Stress in Wheat Seedlings by Modulating Polyamine Metabolism. Front. Plant Sci. 2018, 9, 914. [Google Scholar] [CrossRef] [PubMed]
- Debnath, B.; Islam, W.; Li, M.; Sun, Y.; Lu, X.; Mitra, S.; Hussain, M.; Liu, S.; Qiu, D. Melatonin Mediates Enhancement of Stress Tolerance in Plants. Int. J. Mol. Sci. 2019, 20, 1040. [Google Scholar] [CrossRef]
- Fujiwara, T.; Maisonneuve, S.; Isshiki, M.; Mizutani, M.; Chen, L.; Wong, H.L.; Kawasaki, T.; Shimamoto, K. Sekiguchi Lesion Gene Encodes a Cytochrome P450 Monooxygenase That Catalyzes Conversion of Tryptamine to Serotonin in Rice. J. Biol. Chem. 2010, 285, 11308–11313. [Google Scholar] [CrossRef] [PubMed]
- Hayashi, K.; Fujita, Y.; Ashizawa, T.; Suzuki, F.; Nagamura, Y.; Hayano-Saito, Y. Serotonin attenuates biotic stress and leads to lesion browning caused by a hypersensitive response to Magnaporthe oryzae penetration in rice. Plant J. 2016, 85, 46–56. [Google Scholar] [CrossRef] [PubMed]
- Moustafa-Farag, M.; Almoneafy, A.; Mahmoud, A.; Elkelish, A.; Arnao, M.B.; Li, L.; Ai, S. Melatonin and Its Protective Role against Biotic Stress Impacts on Plants. Biomolecules 2019, 10, 54. [Google Scholar] [CrossRef]
- Shi, H.; Chen, Y.; Tan, D.-X.; Reiter, R.J.; Chan, Z.; He, C. Melatonin induces nitric oxide and the potential mechanisms relate to innate immunity against bacterial pathogen infection in Arabidopsis. J. Pineal Res. 2015, 59, 102–108. [Google Scholar] [CrossRef] [PubMed]
- Zhang, N.; Zhao, B.; Zhang, H.-J.; Weeda, S.; Yang, C.; Yang, Z.-C.; Ren, S.; Guo, Y.-D. Melatonin promotes water-stress tolerance, lateral root formation, and seed germination in cucumber (Cucumis sativus L.). J. Pineal Res. 2012, 54, 15–23. [Google Scholar] [CrossRef] [PubMed]
- Zhang, N.; Zhang, H.-J.; Zhao, B.; Sun, Q.-Q.; Cao, Y.-Y.; Li, R.; Wu, X.-X.; Weeda, S.; Li, L.; Ren, S.; et al. The RNA-seq approach to discriminate gene expression profiles in response to melatonin on cucumber lateral root formation. J. Pineal Res. 2014, 56, 39–50. [Google Scholar] [CrossRef] [PubMed]
- Li, C.; Tan, D.-X.; Liang, D.; Chang, C.; Jia, D.; Ma, F. Melatonin mediates the regulation of ABA metabolism, free-radical scavenging, and stomatal behaviour in two Malus species under drought stress. J. Exp. Bot. 2015, 66, 669–680. [Google Scholar] [CrossRef]
- Li, H.; Dong, Y.; Chang, J.; He, J.; Chen, H.; Liu, Q.; Wei, C.; Ma, J.; Zhang, Y.; Yang, J.; et al. High-throughput microRNA and mRNA sequencing reveals that microRNAs may be involved in melatonin-mediated cold tolerance in Citrulluslanatus L. Front. Plant Sci. 2016, 7, 1231. [Google Scholar]
- Goswami, K.; Mittal, D.; Gautam, B.; Sopory, S.K.; Sanan-Mishra, N. Mapping the salt stress-induced changes in the Root miRNome in Pokkalirice. Biomolecules 2020, 10, 498. [Google Scholar] [CrossRef]
- Balyan, S.; Kumar, M.; Mutum, R.D.; Raghuvanshi, U.; Agarwal, P.; Mathur, S.; Raghuvanshi, S. Identification of miRNA-mediated drought responsive multi-tiered regulatory network in drought tolerant rice, Nagina 22. Sci. Rep. 2017, 7, 15446. [Google Scholar] [CrossRef]
