Intrinsically disordered myelin basic protein (MBP) is one of the key autoantigens in autoimmune neurodegeneration and multiple sclerosis particularly. MBP is highly positively charged and lacks a distinct structure in solution, and therefore its intracellular partners are still mostly enigmatic. Here weauthors used combination of formaldehyde-induced cross-linking followed by immunoprecipitation and liquid chromatography-tandem mass spectrometry (LC-MS/MS) to elucidate the interaction network of MBP in mammalian cells and provide the list of potential MBP interacting proteins. Our data suggest that MBP may be more actively involved in myelination not only as a main building block but also as a self-regulating element.
- myelin basic protein
- MBP
- interactome
- multiple sclerosis
1. Introduction
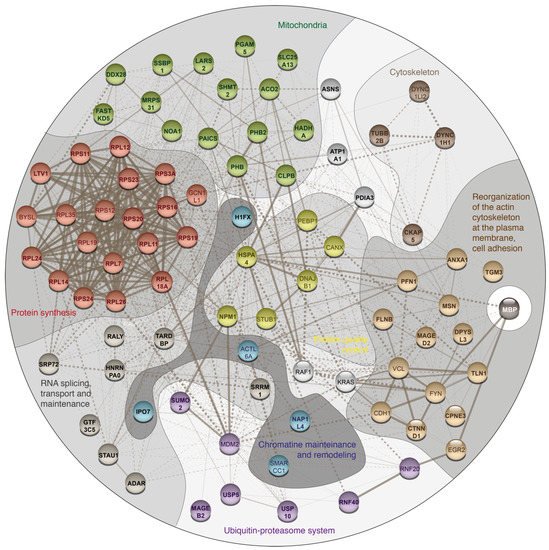
2. Potential Binding Partners of MBP
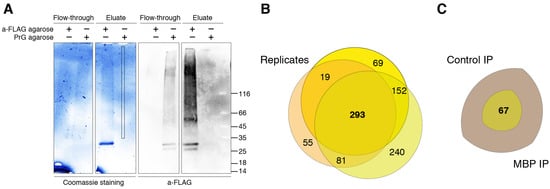
To elucidate potential binding partners of MBP, we performed formaldehyde crosslinking, immunoprecipitation and mass spectrometry analysis of cell lysates of HEK293T cells overexpressing Flag-tagged human MBP (Figure 1). There are several reasons why we used HEK293T cells overexpressing Flag-tagged MBP: (i) There are only a few immortalized cell lines mimicking oligodendrocytes. Despite expression of surface markers similar to those of oligodendrocytes, they still do not fully represent native cells. (ii) Usage of primary oligodendroglial cultures is not obvious due to the heterogenicity and troubles with either transfection or transduction procedures. Finally, amount of endogenous MBP in native oligodendrocytes is dramatically high therefore it may effectively compete with recombinant tagged MBP, displacing it from protein complexes. (iii) Commercially available antibodies partially cross-react with eukaryotic proteome thus generating false-positive results. Additionally, usage of such low-affinity antibodies requires mild washing conditions in contrast to highly specific and affine anti-FLAG antibodies.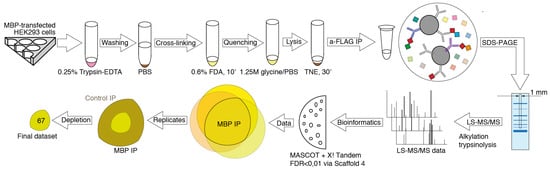
| Protein Name | Gene Name | PSMs for MBP Experiment | PSMs for Control Experiment | ||||
|---|---|---|---|---|---|---|---|
| Protein synthesis | |||||||
| 40S ribosomal protein S16 | RPS16 | 0 | 1 | 2 | 0 | 0 | 0 |
| 40S ribosomal protein S19 | RPS19 | 0 | 1 | 1 | 0 | 0 | 0 |
| 40S ribosomal protein S20 | RPS20 | 0 | 2 | 4 | 0 | 0 | 0 |
| 40S ribosomal protein S23 | RPS23 | 3 | 0 | 4 | 0 | 0 | 0 |
| 60S ribosomal protein L11 | RPL11 | 0 | 1 | 3 | 0 | 0 | 0 |
| 60S ribosomal protein L12 | RPL12 | 0 | 1 | 1 | 0 | 0 | 0 |
| 60S ribosomal protein L18a | RPL18A | 0 | 3 | 8 | 0 | 0 | 0 |
| Protein LTV1 homolog | LTV1 | 0 | 2 | 1 | 0 | 0 | 0 |
| Ribosomal protein L7, isoform CRA_a | RPL7 | 0 | 1 | 4 | 0 | 0 | 0 |
| 40S ribosomal protein S24 | RPS24 | 0 | 2 | 3 | 0 | 0 | 1 |
| 40S ribosomal protein S11 | RPS11 | 0 | 1 | 8 | 0 | 0 | 2 |
| 60S ribosomal protein L26 | RPL26 | 0 | 3 | 6 | 0 | 0 | 2 |
| 40S ribosomal protein S3a | RPS3A | 3 | 2 | 12 | 0 | 0 | 4 |
| 60S ribosomal protein L24 | RPL24 | 0 | 2 | 2 | 0 | 0 | 1 |
| 60S ribosomal protein L14 | RPL14 | 0 | 2 | 5 | 0 | 0 | 2 |
| Mitochondria | |||||||
| 28S ribosomal protein S31, mitochondrial | MRPS31 | 0 | 2 | 2 | 0 | 0 | 0 |
| Aconitate hydratase, mitochondrial | ACO2 | 1 | 0 | 2 | 0 | 0 | 0 |
| cDNA FLJ46863 fis, clone UTERU3011558 | NOA1 | 1 | 0 | 2 | 0 | 0 | 0 |
| FAST kinase domain-containing protein 5, mitochondrial | FASTKD5 | 0 | 2 | 1 | 0 | 0 | 0 |
| Isoform 2 of Caseinolytic peptidase B protein homolog | CLPB | 0 | 2 | 8 | 0 | 0 | 0 |
| Prohibitin (Fragment) | PHB1 | 2 | 2 | 3 | 0 | 0 | 0 |
| Prohibitin-2 | PHB2 | 5 | 2 | 6 | 0 | 0 | 0 |
| Single-stranded DNA-binding protein, mitochondrial (Fragment) | SSBP1 | 1 | 0 | 2 | 0 | 0 | 0 |
| Solute carrier family 25, member 13 (Citrin) variant (Fragment) | SLC25A13 | 0 | 1 | 3 | 0 | 0 | 0 |
| Probable ATP-dependent RNA helicase DDX28 | DDX28 | 0 | 2 | 6 | 0 | 0 | 1 |
| Enoyl-CoA hydratase | HADHA | 3 | 1 | 7 | 0 | 0 | 2 |
| Probable leucine--tRNA ligase, mitochondrial | LARS2 | 0 | 2 | 3 | 0 | 0 | 1 |
| AIR carboxylase (Fragment) | PAICS | 2 | 0 | 6 | 0 | 0 | 2 |
| Serine hydroxymethyltransferase | SHMT2 | 5 | 3 | 17 | 0 | 0 | 7 |
| Serine/threonine-protein phosphatase PGAM5, mitochondrial | PGAM5 | 1 | 0 | 6 | 0 | 0 | 2 |
| mRNA splicing, transport and maintenance | |||||||
| cDNA FLJ77421, highly similar to Homo sapiens autoantigen p542 mRNA | RALY | 2 | 1 | 0 | 0 | 0 | 0 |
| Isoform 2 of General transcription factor 3C polypeptide 5 | GTF3C5 | 0 | 2 | 1 | 0 | 0 | 0 |
| Serine/arginine repetitive matrix 1 isoform 2 (Fragment) | SRRM1 | 0 | 4 | 1 | 0 | 0 | 0 |
| Heterogeneous nuclear ribonucleoprotein A0 | HNRNPA0 | 2 | 2 | 1 | 0 | 0 | 1 |
| Signal recognition particle subunit SRP72 | SRP72 | 1 | 0 | 4 | 0 | 0 | 1 |
| cDNA FLJ75871, highly similar to Homo sapiens staufen, RNA binding protein (STAU), transcript variant T3, mRNA | STAU1 | 0 | 5 | 8 | 0 | 0 | 3 |
| RNA-specific adenosine deaminase | ADAR | 3 | 3 | 1 | 0 | 2 | 0 |
| TAR DNA-binding protein 43 | TARDBP | 4 | 1 | 2 | 0 | 0 | 2 |
| Reorganization of the cytoskeleton and intercellular adhesion | |||||||
| Annexin A1 | ANXA1 | 4 | 3 | 2 | 0 | 0 | 0 |
| Catenin delta-1 | CTNND1 | 0 | 1 | 5 | 0 | 0 | 0 |
| Copine III, isoform CRA_a | CPNE3 | 0 | 2 | 2 | 0 | 0 | 0 |
| Filamin B, beta (Actin binding protein 278), isoform CRA_a | FLNB | 2 | 1 | 4 | 0 | 0 | 0 |
| Moesin | MSN | 2 | 0 | 8 | 0 | 1 | 1 |
| cDNA FLJ56823, highly similar to Protein-glutamine gamma-glutamyltransferase E | TGM3 | 1 | 1 | 2 | 0 | 1 | 0 |
| Profilin-1 | PFN1 | 2 | 1 | 1 | 0 | 0 | 1 |
| Talin-1 | TLN1 | 3 | 4 | 16 | 0 | 1 | 5 |
| Testicular secretory protein Li 7 | DPYSL3 | 0 | 2 | 5 | 0 | 1 | 1 |
| Cytoskeleton and intracellular traffic | |||||||
| Cytoskeleton-associated protein 5 | CKAP5 | 1 | 0 | 8 | 0 | 0 | 1 |
| Cytoplasmic dynein 1 heavy chain 1 | DYNC1H1 | 3 | 0 | 1 | 0 | 0 | 1 |
| Tubulin beta chain | TUBB2B | 0 | 2 | 5 | 0 | 0 | 2 |
| Ubiquitin-proteasome system related | |||||||
| cDNA, FLJ93871, highly similar to Homo sapiens melanoma antigen, family B, 2 (MAGEB2), mRNA | MAGEB2 | 0 | 1 | 3 | 0 | 0 | 0 |
| E3 ubiquitin protein ligase | RNF40 | 0 | 2 | 1 | 0 | 0 | 0 |
| Small ubiquitin-related modifier | SUMO2 | 0 | 2 | 2 | 0 | 0 | 0 |
| Ubiquitin carboxyl-terminal hydrolase | USP10 | 0 | 2 | 1 | 0 | 0 | 0 |
| Melanoma antigen family D, 2, isoform CRA_a | MAGED2 | 0 | 2 | 11 | 0 | 0 | 1 |
| Ubiquitin carboxyl-terminal hydrolase | USP5 | 1 | 1 | 5 | 0 | 0 | 1 |
| Quality control proteins | |||||||
| Heat shock 70 kDa protein 4 | HSPA4 | 2 | 0 | 1 | 0 | 0 | 0 |
| Nucleophosmin (Fragment) | NPM1 | 11 | 3 | 6 | 0 | 0 | 3 |
| Chromatin mainteinance and remodeling | |||||||
| Nucleosome assembly protein 1-like 4, isoform CRA_b | NAP1L4 | 1 | 0 | 3 | 0 | 0 | 0 |
| Histone H1.10 | H1-10 | 2 | 1 | 0 | 0 | 0 | 0 |
| Importin-7 | IPO7 | 0 | 2 | 7 | 0 | 0 | 1 |
| Unassigned to the specific group proteins | |||||||
| Asparagine synthetase [glutamine-hydrolyzing] | ASNS | 0 | 3 | 5 | 0 | 0 | 1 |
| Protein disulfide-isomerase A3 (Fragment) | PDIA3 | 4 | 0 | 1 | 0 | 0 | 1 |
| Sodium/potassium-transporting ATPase subunit alpha (Fragment) | ATP1A1 | 0 | 1 | 3 | 0 | 0 | 1 |
| BAF53A protein | BAF53A | 2 | 0 | 1 | 0 | 0 | 0 |
| Peroxisome proliferator activated receptor interacting complex protein | PRIC295 | 1 | 0 | 9 | 0 | 0 | 1 |
References
- Benjamins, J.A.; Morell, P. Proteins of Myelin and Their Metabolism. Neurochem. Res. 1978, 3, 137–174.
- Garbay, B.; Fournier, M.; Sallafranque, M.L.; Muller, S.; Boiron, F.; Heape, A.; Cassagne, C.; Bonnet, J. Po, MBP, Histone, and DNA Levels in Sciatic Nerve. Neurochem. Pathol. 1988, 8, 91–107.
- Krigbaum, W.R.; Hsu, T.S. Molecular Conformation of Bovine A1 Basic Protein, a Coiling Macromolecule in Aqueous Solution. Biochemistry 1975, 14, 2542–2546.
- Polverini, E.; Fasano, A.; Zito, F.; Riccio, P.; Cavatorta, P. Conformation of Bovine Myelin Basic Protein Purified with Bound Lipids. Eur. Biophys. J. EBJ 1999, 28, 351–355.
- Harauz, G.; Ishiyama, N.; Hill, C.M.D.; Bates, I.R.; Libich, D.S.; Farès, C. Myelin Basic Protein-Diverse Conformational States of an Intrinsically Unstructured Protein and Its Roles in Myelin Assembly and Multiple Sclerosis. Micron 2004, 35, 503–542.
- Harauz, G.; Libich, D. The Classic Basic Protein of Myelin—Conserved Structural Motifs and the Dynamic Molecular Barcode Involved in Membrane Adhesion and Protein-Protein Interactions. Curr. Protein Pept. Sci. 2009, 10, 196–215.
- Aggarwal, S.; Snaidero, N.; Pähler, G.; Frey, S.; Sánchez, P.; Zweckstetter, M.; Janshoff, A.; Schneider, A.; Weil, M.-T.; Schaap, I.A.T.; et al. Myelin Membrane Assembly Is Driven by a Phase Transition of Myelin Basic Proteins Into a Cohesive Protein Meshwork. PLoS Biol. 2013, 11, e1001577.
- Boggs, J.M. Myelin Basic Protein: A Multifunctional Protein. Cell. Mol. Life Sci. CMLS 2006, 63, 1945–1961.
- Boggs, J.M.; Yip, P.M.; Rangaraj, G.; Jo, E. Effect of Posttranslational Modifications to Myelin Basic Protein on Its Ability to Aggregate Acidic Lipid Vesicles. Biochemistry 1997, 36, 5065–5071.
- Bates, I.R.; Boggs, J.M.; Feix, J.B.; Harauz, G. Membrane-Anchoring and Charge Effects in the Interaction of Myelin Basic Protein with Lipid Bilayers Studied by Site-Directed Spin Labeling. J. Biol. Chem. 2003, 278, 29041–29047.
- Hill, C.M.D.; Harauz, G. Charge Effects Modulate Actin Assembly by Classic Myelin Basic Protein Isoforms. Biochem. Biophys. Res. Commun. 2005, 329, 362–369.
- Homchaudhuri, L.; Polverini, E.; Gao, W.; Harauz, G.; Boggs, J.M. Influence of Membrane Surface Charge and Post-Translational Modifications to Myelin Basic Protein on Its Ability to Tether the Fyn-SH3 Domain to a Membrane in Vitro. Biochemistry 2009, 48, 2385–2393.
- Belogurov, A.; Kudriaeva, A.; Kuzina, E.; Smirnov, I.; Bobik, T.; Ponomarenko, N.; Kravtsova-Ivantsiv, Y.; Ciechanover, A.; Gabibov, A. Multiple Sclerosis Autoantigen Myelin Basic Protein Escapes Control by Ubiquitination during Proteasomal Degradation. J. Biol. Chem. 2014, 289, 17758–17766.
- Kudriaeva, A.; Kuzina, E.S.; Zubenko, O.; Smirnov, I.V.; Belogurov, A. Charge-Mediated Proteasome Targeting. FASEB J. 2019, 33, 6852–6866.
- Lutton, J.D.; Winston, R.; Rodman, T.C. Multiple Sclerosis: Etiological Mechanisms and Future Directions. Exp. Biol. Med. 2004, 229, 12–20.
- Sospedra, M.; Martin, R. Immunology of Multiple Sclerosis. Annu. Rev. Immunol. 2005, 23, 683–747.
- Belogurov, A.A.; Kurkova, I.N.; Friboulet, A.; Thomas, D.; Misikov, V.K.; Zakharova, M.Y.; Suchkov, S.V.; Kotov, S.V.; Alehin, A.I.; Avalle, B.; et al. Recognition and Degradation of Myelin Basic Protein Peptides by Serum Autoantibodies: Novel Biomarker for Multiple Sclerosis. J. Immunol. 1950 2008, 180, 1258–1267.
- Ponomarenko, N.A.; Durova, O.M.; Vorobiev, I.I.; Belogurov, A.A.; Kurkova, I.N.; Petrenko, A.G.; Telegin, G.B.; Suchkov, S.V.; Kiselev, S.L.; Lagarkova, M.A.; et al. Autoantibodies to Myelin Basic Protein Catalyze Site-Specific Degradation of Their Antigen. Proc. Natl. Acad. Sci. USA 2006, 103, 281–286.
- Ponomarenko, N.A.; Durova, O.M.; Vorobiev, I.I.; Belogurov, A.A.; Telegin, G.B.; Suchkov, S.V.; Misikov, V.K.; Morse, H.C.; Gabibov, A.G. Catalytic Activity of Autoantibodies toward Myelin Basic Protein Correlates with the Scores on the Multiple Sclerosis Expanded Disability Status Scale. Immunol. Lett. 2006, 103, 45–50.
- Kuzina, E.S.; Chernolovskaya, E.L.; Kudriaeva, A.A.; Zenkova, M.A.; Knorre, V.D.; Surina, E.A.; Ponomarenko, N.A.; Bobik, T.V.; Smirnov, I.V.; Bacheva, A.V.; et al. Immunoproteasome Enhances Intracellular Proteolysis of Myelin Basic Protein. Dokl. Biochem. Biophys. 2013, 453, 300–303.
- Kim, J.K.; Mastronardi, F.G.; Wood, D.D.; Lubman, D.M.; Zand, R.; Moscarello, M.A. Multiple Sclerosis: An Important Role for Post-Translational Modifications of Myelin Basic Protein in Pathogenesis. Mol. Cell. Proteom. 2003, 2, 453–462.
- Kuzina, E.S.; Kudriaeva, A.A.; Glagoleva, I.S.; Knorre, V.D.; Gabibov, A.G.; Belogurov, A.A. Deimination of the Myelin Basic Protein Decelerates Its Proteasome-Mediated Metabolism. Dokl. Biochem. Biophys. 2016, 469, 277–280.
- Harauz, G.; Musse, A.A. A Tale of Two Citrullines—Structural and Functional Aspects of Myelin Basic Protein Deimination in Health and Disease. Neurochem. Res. 2007, 32, 137–158.
- Wang, C.; Neugebauer, U.; Bürck, J.; Myllykoski, M.; Baumgärtel, P.; Popp, J.; Kursula, P. Charge Isomers of Myelin Basic Protein: Structure and Interactions with Membranes, Nucleotide Analogues, and Calmodulin. PLoS ONE 2011, 6, e19915.
- Kuzina, E.; Kudriaeva, A.; Smirnov, I.; Dubina, M.V.; Gabibov, A.; Belogurov, A. Glatiramer Acetate and Nanny Proteins Restrict Access of the Multiple Sclerosis Autoantigen Myelin Basic Protein to the 26S Proteasome. BioMed Res. Int. 2014, 2014, 926394.
- Belogurov, A.; Kuzina, E.; Kudriaeva, A.; Kononikhin, A.; Kovalchuk, S.; Surina, Y.; Smirnov, I.; Lomakin, Y.; Bacheva, A.; Stepanov, A.; et al. Ubiquitin-Independent Proteosomal Degradation of Myelin Basic Protein Contributes to Development of Neurodegenerative Autoimmunity. FASEB J. 2015, 29, 1901–1913.
- Sutherland, B.W.; Toews, J.; Kast, J. Utility of Formaldehyde Cross-Linking and Mass Spectrometry in the Study of Protein–Protein Interactions. J. Mass Spectrom. 2008, 43, 699–715.
- Klockenbusch, C.; Kast, J. Optimization of Formaldehyde Cross-Linking for Protein Interaction Analysis of Non-Tagged Integrin β 1. J. Biomed. Biotechnol. 2010, 2010, 1–13.
- Szklarczyk, D.; Gable, A.L.; Nastou, K.C.; Lyon, D.; Kirsch, R.; Pyysalo, S.; Doncheva, N.T.; Legeay, M.; Fang, T.; Bork, P.; et al. The STRING Database in 2021: Customizable Protein-Protein Networks, and Functional Characterization of User-Uploaded Gene/Measurement Sets. Nucleic Acids Res. 2021, 49, D605–D612.
- Szklarczyk, D.; Gable, A.L.; Lyon, D.; Junge, A.; Wyder, S.; Huerta-Cepas, J.; Simonovic, M.; Doncheva, N.T.; Morris, J.H.; Bork, P.; et al. STRING V11: Protein-Protein Association Networks with Increased Coverage, Supporting Functional Discovery in Genome-Wide Experimental Datasets. Nucleic Acids Res. 2019, 47, D607–D613.
