Cultivated cardoon (Cynara cardunculus var. altilis L.) is a promising candidate species for the development of plant cell cultures (PCC) suitable for large-scale biomass production and recovery of nutraceuticals. We set up the first successful protocol for the stable genetic transformation via Agrobacterium tumefaciens of cardoon PCC, a valuable achievement for the improvement of their biorefinery potential.
- plant cell cultures
- lignin
- cellulose accessibility
- nutraceuticals
- biorefinery
- MYB4
1. Introduction
2. Development of a Method for the Stable Transformation of Cardoon
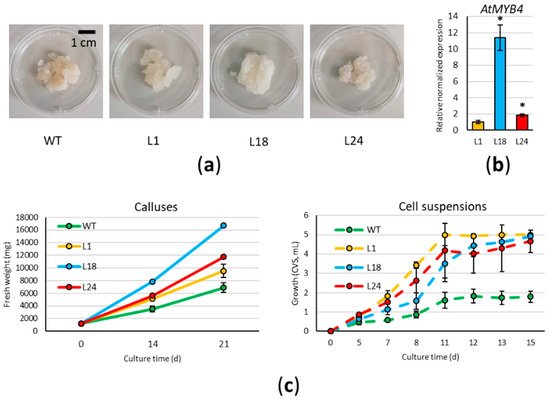
3. AtMYB4oe Lines Have Higher Growth Rates Than Wild Type
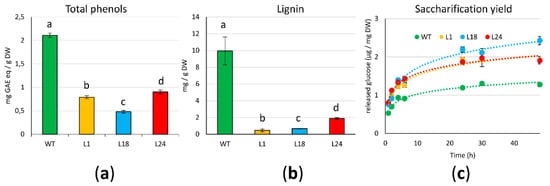
4. AtMYB4oe Lines Show Decreased Phenolic Compounds and Lignin Content and Enhanced Enzymatic Saccharification Efficiency
5. Transcriptomic analysis
6. Biochemical profiles
WT and transgenic lines were analyzed for the characterization of phenolic compounds, antioxidant activities, and fatty acids (Table 1). This deep quantitative and qualitative characterization revealed interesting findings about specific nutraceuticals: CGA accumulates more than WT. Similarly, p-coumaric acid and flavonoids identified in the extracts (quercetin, kaempferol, naringin, luteolin, myricetin, and apigenin) accumulated at similar levels than WT. Notably, this confirms that while the overexpression of AtMYB4 represses the phenylpropanoid pathway by reducing the production of lignin monomers, it does not necessarily depauperate nutraceuticals, which could be recovered from the cell cultures, increasing the interest in these lines from a biotechnological point of view. Interestingly, we detected a 3–4-fold increase in L1 and L24 lines for oleic acid, whose increase in leves represents a goal of biorefinery approaches in many plant species.
WT |
L1 |
L18 |
L24 |
||
|---|---|---|---|---|---|
Polyphenols (µg/g DW) |
5-iFQA | 21.50 ± 1.33a | 15.41 ± 0.43b | 0.11 ± 0.04c | 4.26 ± 0.12d |
| 1,5-DiCQA (cynarin) | 3345.79 ± 112.23a | 532.70 ± 11.55b | 2.50 ± 0.81c | 352.98 ± 21.71d | |
| 3,4-DiCQA | 3152.41 ± 26.67a | 443.88 ± 12.43b | 103.58 ± 8.23c | 1615.65 ± 34.56d | |
| 5-FQA | 98.87 ± 11.34a | 5.56 ± 0.43b | 0.83 ± 0.011c | 76.44 ± 11.34d | |
| 3-FQA | 3.09 ± 0.55a | 2.71 ± 0.91b | 0.10 ± 0.01c | 10.20 ± 0.23d | |
| 3-CQA (CGA) | 319.38 ± 22.45a | 424.66 ± 11.32b | 97.28 ± 3.56c | 513.48 ± 13.57d | |
| p-coumaric acid | 3.50 ± 0.02a | 3.75 ± 0.91a | 1.50 ± 0.01b | 3.85 ± 0.02a | |
| Quercetin | 3.35 ± 0.02a | 4.30 ± 0.02b | 0.55 ± 0.01c | 4.35 ± 0.02b | |
| Quercetin-glucoside | 4.00 ± 0.65a | 5.25 ± 1.23b | 1.25 ± 0.03c | 5.40 ± 0.34b | |
| Kaempferol | 3.55 ± 0.65a | 4.30 ± 0.91b | 1.05 ± 0.03c | 4.55 ± 0.83d | |
| Kaempferol-3-O-glucoside | 1.15 ± 0.02a | 1.50 ± 0.03b | 0.40 ± 0.04c | 1.60 ± 0.04b | |
| Naringin | 1.20 ± 0.01a | 1.75 ± 0.01b | 0.41 ± 0.03c | 1.800.03b | |
| Luteolin | 1.50 ± 0.03a | 2.35 ± 0.02b | 0.60 ± 0.04c | 2.55 ± 0.04d | |
| Myricetin | 2.55 ± 0.34a | 2.96 ± 0.32b | 0.73 ± 0.11c | 3.04 ± 0.12b | |
| Apigenin | 0.20 ± 0.03a | 0.25 ± 0.02b | 0.01 ± 0.003c | 0.30 ± 0.01d | |
| Total polyphenols | 6962.05a | 1451.33b | 210.92c | 2600.46d | |
Antiox. activity (TEAC) |
DPPH | 83.7 ± 0.61a | 22.17 ± 0.42b | 16.96 ± 0.63c | 35.43 ± 5.82d |
| ABTS | 71.33 ± 0.20a | 19.16 ± 0.07b | 12.54 ± 0.26c | 23.61 ± 0.22d | |
| FRAP | 66.81 ± 0.98a | 30.84 ± 0.21b | 11.09 ± 0.08c | 27.90 ± 0.54d | |
Oil % |
11.38 | 8.24 | 10.35 | 7.25 | |
Fatty acids (%) |
Palmitic (C16:0) | 22.29 ± 1.12ab | 21.71 ± 0.58a | 24.06 ± 0.35b | 19.46 ± 0.38c |
| Stearic (C18:0) | 2.85 ± 0.31ab | 3.14 ± 0.54ab | 2.29 ± 0.07a | 3.73 ± 0.26b | |
| Oleic (C18:1) | 3.43 ± 1.91a | 10.30 ± 0.56b | 3.49 ± 0.27a | 12.35 ± 0.16c | |
| Linoleic (C18:2) | 19.38 ± 3.74a | 45.82 ± 0.85b | 41.04 ± 0.61c | 41.34 ± 0.39c | |
| Linolenic (C18:3) | 42.60 ± 6.34a | 10.16 ± 0.38b | 20.73 ± 0.74c | 14.46 ± 0.06d | |
| Arachidic (C20:0) | 0.80 ± 0.08a | 0.88 ± 0.01a | 0.84 ± 0.05a | 1.04 ± 0.05b | |
| Lignoceric (C24:0) | 2.42 ± 0.30a | 3.26 ± 1.59a | 2.41 ± 0.58a | 2.40 ± 0.68a | |
| Nervonic (C24:1) | 1.23 ± 0.14a | 0.18 ± 0.03b | 0.52 ± 0.47ab | 0.66 ± 0.12ab | |
| Total SFA% | 30.54 ± 0.78ab | 30.29 ± 0.76a | 32.07 ± 0.43b | 28.00 ± 0.10c | |
| Total MUFA % | 4.65 ± 1.777a | 10.48 ± 0.30b | 4.02 ± 0.21a | 13.01 ± 0.28c | |
| Total PUFA % | 61.98 ± 2.88a | 55.98 ± 0.80b | 61.77 ± 0.41a | 55.81 ± 0.42b | |
| Others % | 2.83 | 3.25 | 2.14 | 3.18 |
7. Conclusions and future perspectives
We have shown that the protocol we set up allows the stable genetic transformation of cardoon cells. The AtMYB4oe lines presented in this study proved to be valuable tools for their use in bioreactors. The main advantages of these lines are represented by their faster growth rate and improved accessibility of the biomass to enzymatic degradation, due to the reduction in lignin content, which also implies an easier extractability of compounds of interest, as well as an interesting modification in their nutraceutical value. The development of this technique represents a significant step towards the industrial use of cardoon cell cultures, which can be further improved targeting specific metabolic pathways of interest; two examples would be represented by targeting other MYB transcription factors involved in the production of specialized metabolites, or by the alteration of the activity of biosynthetic genes for fatty acids, both in the frame of gain or loss-of-function genetic approaches. Moreover, further exploration of the generated RNA-seq data could provide useful to further support molecular analyses of primary and specialized metabolic pathways of cardoon cell cultures. Finally, in order to evaluate whether the use of cardoon cells for biorefinery is energetically and economically sustainable, further studies on large-scale production are being conducted.
References
- Barracosa, P.; Barracosa, M.; Pires, E. Cardoon as a Sustainable Crop for Biomass and Bioactive Compounds Production. Chem. Biodivers. 2019, 16, e1900498.
- Basnizki, J.; Zohary, D. Breeding of Seed-Planted Artichoke. In Plant Breeding Reviews; John Wiley & Sons, Inc.: Oxford, UK, 2010; pp. 253–269.
- Acquadro, A.; Barchi, L.; Portis, E.; Mangino, G.; Valentino, D.; Mauromicale, G.; Lanteri, S. Genome reconstruction in Cynara cardunculus taxa gains access to chromosome-scale DNA variation. Sci. Rep. 2017, 7, 5617.
- Almeida, C.M.; Simões, I. Cardoon-based rennets for cheese production. Appl. Microbiol. Biotechnol. 2018, 102, 4675–4686.
- Toscano, V.; Sollima, L.; Genovese, C.; Melilli, M.G.; Raccuia, S.A. Pilot plant system for biodiesel and pellet production from cardoon: Technical and economic feasibility. In Proceedings of the Acta Horticulturae, International Society for Horticultural Science, La Plata, Argentina, 17 November 2016; Volume 1147, pp. 429–442.
- Pappalardo, H.D.; Toscano, V.; Puglia, G.D.; Genovese, C.; Raccuia, S.A. Cynara cardunculus L. as a Multipurpose Crop for Plant Secondary Metabolites Production in Marginal Stressed Lands. Front. Plant Sci. 2020, 11, 240.
- Gominho, J.; Curt, M.D.; Lourenço, A.; Fernández, J.; Pereira, H. Cynara cardunculus L. as a biomass and multi-purpose crop: A review of 30 years of research. Biomass Bioenergy 2018, 109, 257–275.
- Colla, G.; Rouphael, Y.; Cardarelli, M.; Svecova, E.; Rea, E.; Lucini, L. Effects of saline stress on mineral composition, phenolic acids and flavonoids in leaves of artichoke and cardoon genotypes grown in floating system. J. Sci. Food Agric. 2013, 93, 1119–1127.
- Docimo, T.; De Stefano, R.; Cappetta, E.; Piccinelli, A.L.; Celano, R.; De Palma, M.; Tucci, M. Physiological, Biochemical, and Metabolic Responses to Short and Prolonged Saline Stress in Two Cultivated Cardoon Genotypes. Plants 2020, 9, 554.
- Gominho, J.; Lourenço, A.; Palma, P.; Lourenço, M.E.; Curt, M.D.; Fernández, J.; Pereira, H. Large scale cultivation of Cynara cardunculus L. for biomass production—A case study. Ind. Crop. Prod. 2011, 33, 1–6.
- Sorrentino, M.C.; Capozzi, F.; Amitrano, C.; Giordano, S.; Arena, C.; Spagnuolo, V. Performance of three cardoon cultivars in an industrial heavy metal-contaminated soil: Effects on morphology, cytology and photosynthesis. J. Hazard. Mater. 2018, 351, 131–137.
- Capozzi, F.; Sorrentino, M.C.; Caporale, A.G.; Fiorentino, N.; Giordano, S.; Spagnuolo, V. Exploring the phytoremediation potential of Cynara cardunculus: A trial on an industrial soil highly contaminated by heavy metals. Environ. Sci. Pollut. Res. 2020, 27, 9075–9084.
- Piscioneri, I.; Sharma, N.; Baviello, G.; Orlandini, S. Promising industrial energy crop, Cynara cardunculus: A potential source for biomass production and alternative energy. Energy Convers. Manag. 2000, 41, 1091–1105.
- Fernández, J.; Curt, M.D.; Aguado, P.L. Industrial applications of Cynara cardunculus L. for energy and other uses. Ind. Crop. Prod. 2006, 24, 222–229.
- Mantineo, M.; D’Agosta, G.M.; Copani, V.; Patanè, C.; Cosentino, S.L. Biomass yield and energy balance of three perennial crops for energy use in the semi-arid Mediterranean environment. Field Crop. Res. 2009, 114, 204–213.
- Foti, S.; Mauromicale, G.; Raccuia, S.A.; Fallico, B.; Fanella, F.; Maccarone, E. Possible alternative utilization of Cynara spp. I. Biomass, grain yield and chemical composition of grain. Ind. Crop. Prod. 1999, 10, 219–228.
- Ierna, A.; Sortino, O.; Mauromicale, G. Biomass, seed and energy yield of Cynara cardunculus L. as affected by environment and season. Agronomy 2020, 10, 1584.
- Gominho, J.; Fernandez, J.; Pereira, H. Cynara cardunculus L. A new fibre crop for pulp and paper production. Ind. Crop. Prod. 2001, 13, 1–10.
- Barbosa, C.H.; Andrade, M.A.; Vilarinho, F.; Castanheira, I.; Fernando, A.L.; Loizzo, M.R.; Silva, A.S. A new insight on cardoon: Exploring new uses besides cheese making with a view to zero waste. Foods 2020, 9, 564.
- Pandino, G.; Lombardo, S.; Mauromicale, G.; Williamson, G. Phenolic acids and flavonoids in leaf and floral stem of cultivated and wild Cynara cardunculus L. genotypes. Food Chem. 2011, 126, 417–422.
- Pinelli, P.; Agostini, F.; Comino, C.; Lanteri, S.; Portis, E.; Romani, A. Simultaneous quantification of caffeoyl esters and flavonoids in wild and cultivated cardoon leaves. Food Chem. 2007, 105, 1695–1701.
- Graziani, G.; Docimo, T.; De Palma, M.; Sparvoli, F.; Izzo, L.; Tucci, M.; Ritieni, A. Changes in Phenolics and Fatty Acids Composition and Related Gene Expression during the Development from Seed to Leaves of Three Cultivated Cardoon Genotypes. Antioxidants 2020, 9, 1096.
- Kollia, E.; Markaki, P.; Zoumpoulakis, P.; Proestos, C. Antioxidant activity of Cynara scolymus L. and Cynara cardunculus L. extracts obtained by different extraction techniques. Nat. Prod. Res. 2017, 31, 1163–1167.
- Falleh, H.; Ksouri, R.; Chaieb, K.; Karray-Bouraoui, N.; Trabelsi, N.; Boulaaba, M.; Abdelly, C. Phenolic composition of Cynara cardunculus L. organs, and their biological activities. Comptes Rendus Biol. 2008, 331, 372–379.
- Naveed, M.; Hejazi, V.; Abbas, M.; Kamboh, A.A.; Khan, G.J.; Shumzaid, M.; Ahmad, F.; Babazadeh, D.; FangFang, X.; Modarresi-Ghazani, F.; et al. Chlorogenic acid (CGA): A pharmacological review and call for further research. Biomed. Pharmacother. 2018, 97, 67–74.
- Mehmetçik, G.; Özdemirler, G.; Koçak-Toker, N.; Çevikbaş, U.; Uysal, M. Effect of pretreatment with artichoke extract on carbon tetrachloride-induced liver injury and oxidative stress. Exp. Toxicol. Pathol. 2008, 60, 475–480.
- Küçükgergin, C.; Aydın, A.F.; Özdemirler-Erata, G.; Mehmetçik, G.; Koçak-Toker, N.; Uysal, M. Effect of artichoke leaf extract on hepatic and cardiac oxidative stress in rats fed on high cholesterol diet. Biol. Trace Elem. Res. 2010, 135, 264–274.
- Kukić, J.; Popović, V.; Petrović, S.; Mucaji, P.; Ćirić, A.; Stojković, D.; Soković, M. Antioxidant and antimicrobial activity of Cynara cardunculus extracts. Food Chem. 2008, 107, 861–868.
- Zoghlami, A.; Paës, G. Lignocellulosic Biomass: Understanding Recalcitrance and Predicting Hydrolysis. Front. Chem. 2019, 7, 874.
- Kumar, A.K.; Sharma, S. Recent updates on different methods of pretreatment of lignocellulosic feedstocks: A review. Bioresour. Bioprocess. 2017, 4, 7.
- Halpin, C. Lignin engineering to improve saccharification and digestibility in grasses. Curr. Opin. Biotechnol. 2019, 56, 223–229.
- Maccarone, E.; Fallico, B.; Fanella, F.; Mauromicale, G.; Raccuia, S.A.; Foti, S. Possible alternative utilization of Cynara spp. II. Chemical characterization of their grain oil. Ind. Crop. Prod. 1999, 10, 229–237.
- Lavermicocca, P.; Rossi, M.; Russo, F.; Srirajaskanthan, R. Chapter 77—Table Olives: A Carrier for Delivering Probiotic Bacteria to Humans A2—Preedy, Victor R; Academic Press: New York, NY, USA, 2010; pp. 735–743.
- Wellenreuther, C.; Wolf, A. Innovative Feedstocks in Biodegradable Bio-Based Plastics: A Literature Review; Hamburgisches Welt-Wirtschafts-Archiv (HWWA): Hamburg, Germany, 2020.
- Koh, M.Y.; Tinia, T.I. A review of biodiesel production from Jatropha curcas L. oil. Renew. Sustain. Energy Rev. 2011, 15, 2240–2251.
- Metzger, J.O.; Bornscheuer, U. Lipids as renewable resources: Current state of chemical and biotechnological conversion and diversification. Appl. Microbiol. Biotechnol. 2006, 71, 13–22.
- Ierna, A.; Mauro, R.P.; Mauromicale, G. Biomass, grain and energy yield in Cynara cardunculus L. as affected by fertilization, genotype and harvest time. Biomass Bioenergy 2012, 36, 404–410.
- Ottaiano, L.; Di Mola, I.; Impagliazzo, A.; Cozzolino, E.; Masucci, F.; Mori, M.; Fagnano, M. Yields and quality of biomasses and grain in Cynara cardunculus L. grown in southern Italy, as affected by genotype and environmental conditions. Ital. J. Agron. 2017, 12, 375–382.
- Pandino, G.; Lombardo, S.; Moglia, A.; Portis, E.; Lanteri, S.; Mauromicale, G. Leaf polyphenol profile and SSR-based fingerprinting of new segregant Cynara cardunculus genotypes. Front. Plant Sci. 2015, 5, 800.
- Sharma, A.; Shahzad, B.; Rehman, A.; Bhardwaj, R.; Landi, M.; Zheng, B. Response of phenylpropanoid pathway and the role of polyphenols in plants under abiotic stress. Molecules 2019, 24, 2452.
- Curt, M.D.; Sánchez, G.; Fernández, J. The potential of Cynara cardunculus L. for seed oil production in a perennial cultivation system. Biomass Bioenergy 2002, 23, 33–46.
- Raccuia, S.A.; Piscioneri, I.; Sharma, N.; Melilli, M.G. Genetic variability in Cynara cardunculus L. domestic and wild types for grain oil production and fatty acids composition. Biomass Bioenergy 2011, 35, 3167–3173.
- Balestrini, R.; Brunetti, C.; Cammareri, M.; Caretto, S.; Cavallaro, V.; Cominelli, E.; De Palma, M.; Docimo, T.; Giovinazzo, G.; Grandillo, S.; et al. Strategies to Modulate Specialized Metabolism in Mediterranean Crops: From Molecular Aspects to Field. Int. J. Mol. Sci. 2021, 22, 2887.
- Nielsen, E.; Temporiti, M.E.E.; Cella, R. Improvement of phytochemical production by plant cells and organ culture and by genetic engineering. Plant Cell Rep. 2019, 38, 1199–1215.
- Ferid, A.; Mohammed, A.; Khalivulla, S.I.; Korivi, M.; Abdul Razab, M.K.A. Plant Cell and Callus Cultures as an Alternative Source of Bioactive Compounds with Therapeutic Potential against Coronavirus Disease (COVID-19). IOP Conf. Ser. Earth Environ. Sci. 2020, 596, 012099.
- Ramachandra Rao, S.; Ravishankar, G.A. Plant cell cultures: Chemical factories of secondary metabolites. Biotechnol. Adv. 2002, 20, 101–153.
- Efferth, T. Biotechnology Applications of Plant Callus Cultures. Engineering 2019, 5, 50–59.
- Wang, X.C.; Wu, J.; Guan, M.L.; Zhao, C.H.; Geng, P.; Zhao, Q. Arabidopsis MYB4 plays dual roles in flavonoid biosynthesis. Plant J. 2020, 101, 637–652.
