Alzheimer’s disease (AD) is the most frequently diagnosed neurodegenerative disorder worldwide and the sixth leading cause of death in the United States and is on the rise [1]. The disorder is characterised by amyloid plaques and neurofibrillary tangles; cell loss, vascular damage and dementia follow as a direct result of the vicious cycle of their deposition. Along with age and family history, inheritance also plays an essential role in the development of AD. Among several genetic risk factors, the APP, PSEN1, PSEN2 had been identified as a causative factor for early-onset AD, the APOE-ε4 allele (encodes a protein that transports cholesterol in the bloodstream) was shown to have a strong impact on late-onset AD. However, studies based on APOE status among different racial and ethnic groups have shown inconsistent results .Despite a number of treatments being approved by the U.S. Food and Drug Administration (FDA), none of those therapeutic strategies can cure the disease . There is strong evidence that early diagnosis and treatment might help to decelerate the progression of the disease and maintain brain function. Therefore, rational development of medical approaches (e.g., sophisticated brain imaging studies and discovery of novel candidate genes, proteins, and other substances in blood or cerebrospinal fluid) are fundamental for better understanding of the molecular factors that contribute to disease progression, and for improving the early diagnosis and treatment decisions.
- Alzheimer’s disease
- gene co-expression network
- genome-scale metabolic model
1. Distinct Gene Expression and Functional Profiles in Different Brain Regions of AD
| VS | p-Val (Hyper) | Overlap | PathwayRegion_1 DEG Size |
Region_2 DEG Size |
Both CodingGene Size | ||||
|---|---|---|---|---|---|---|---|---|---|
| Shared DEGs in the Pathway | |||||||||
| RO-MT | 0.00054 | 128 | 2885 | 477 | 14,001 | ||||
| hsa03450_Non-homologous_end-joining | NHEJ1 | MC-RO | 0.00516 | 347 | 2885 | 1515 | 14,186 | ||
| MT-MC | 9.20599 × 10−11 | 98 | 477 | 1515 | 14,186 | ||||
| Gene Symbol | Associated Protein Name (UNIPROT) | Most Associated KEGG Pathways | |||||
|---|---|---|---|---|---|---|---|
| ADAMTS2 ¥ | A disintegrin and metalloproteinase with thrombospondin motifs 2 | - | |||||
| ATP1B3 # | Sodium/potassium-transporting ATPase subunit beta-3 | Secretion (insulin, salivary, bile, gastric acid, pancreatic, aldosterone) has04961_Endocrine_and_other_factor-regulated_calcium_reabsorption has04973_Carbohydrate_digestion_and_absorption has04260_Cardiac_muscle_contraction has04978_Mineral_absorption |
|||||
| hsa04010_MAPK_signaling_pathway | DAXX | RAF1 | BTG # | B-cell translocation gene 1 protein | has03018_RNA_degradation | ||
| hsa04012_ErbB_signaling_pathway | RAF1 | STAT5B | DAXX # | Death domain-associated protein 6 | has04010_MAPK_signaling_pathway, has04210_Apoptosis, has05012_Parkinson_disease, has05014_Amyotrophic_lateral_sclerosis, has05022_Pathways_of_neurodegeneration, has05168_Herpes_simplex_virus_1_infection | ||
| hsa04014_Ras_signaling_pathway | RAF1 | RALBP1 | DYNC2LI1 * | Cytoplasmic dynein 2 light intermediate chain 1 | has04962_Vasopressin-regulated_water_reabsorption, has05132_Salmonella_infection | ||
| hsa04015_Rap1_signaling_pathway | RAF1 | FAM129B # | Protein Niban 2 | - | |||
| FAM167B # | Protein FAM167B | - | |||||
| FAM90A1 * | Protein FAM90A1 | - | |||||
| FBXO2 # | F-box only protein 2 | has04068_FoxO_signaling_pathway, has04120_Ubiquitin_mediated_proteolysis, has04141_Protein_processing_in_endoplasmic_reticulum, has05132_Salmonella_infection |
|||||
| GARNL3 # | GTPase-activating Rap/Ran-GAP domain-like protein 3 | - | |||||
| GIT1 # | ARF GTPase-activating protein GIT1 | has04144_Endocytosis, has04810_Regulation_of_actin_cytoskeleton, has05120_Epithelial_cell_signaling_in_Helicobacter_pylori_infection |
|||||
| H2AFV # | Histone H2A.V | - | |||||
| HEBP2 # | Heme-binding protein 2 | - | |||||
| ID3 # | DNA-binding protein inhibitor ID-3 | has04350_TGF-beta_signaling_pathway | |||||
| NHEJ1 # | Non-homologous end-joining factor 1 | has03450_Non-homologous_end-joining | |||||
| NT5DC2 # | 5’-nucleotidase domain-containing protein 2 | has04935_Growth_hormone_synthesis, secretion_and_action has04726_Serotonergic_synapse, has04510_Focal_adhesion |
|||||
| RALBP1 # | RalBP1-associated Eps domain-containing protein 2 | has04014_Ras_signaling_pathway, has05212_Pancreatic_cancer | |||||
| - | |||||||
| PAFAH1B3 # | Platelet-activating factor acetylhydrolase IB subunit alpha1 | S100A4 # | Calvasculin/Metastasin | - | |||
| S100A6 # | Calcyclin/Growth factor-inducible protein 2A9 | - | |||||
| SEPT9 # | Septin-9 | - | |||||
| SMAD4 # | Mothers against decapentaplegic homolog 4 | Signalling (Fox0, Wnt, apelin etc.), Cancer (colorectal etc.) has04110_Cell_cycle, has04520_Adherens_junction |
|||||
| STARD10 # | START domain-containing protein 10 | - | |||||
| STAT5B # | Signal transducer and activator of transcription 5B | Signalling (AGE-RAGE, JAK/STAT etc.), Myeloid leukemia has04217_Necroptosis, has04659_Th17_cell_differentiation |
|||||
| TRIM45 * | Tripartite motif-containing protein 45 | - | |||||
| TRIM66 * | Tripartite motif-containing protein 66 | - | |||||
| TRIP10 | |||||||
| hsa04022_cGMP-PKG_signaling_pathway | ATP1B3 | RAF1 | |||||
| hsa04024_cAMP_signaling_pathway | ATP1B3 | RAF1 | |||||
| hsa04062_Chemokine_signaling_pathway | RAF1 | STAT5B | |||||
| hsa04261_Adrenergic_signaling_in_cardiomyocytes | ATP1B3 | ||||||
| hsa04210_Apoptosis | DAXX | RAF1 | has00565_Ether_lipid_metabolism | ||||
| hsa04310_Wnt_signaling_pathway | SMAD4 | ||||||
| hsa04350_TGF-beta_signaling_pathway | ID3 | SMAD4 | PLAGL1 * | Zinc finger protein PLAGL1 | - | ||
| hsa04370_VEGF_signaling_pathway | RAF1 | RAF1 # | RAF proto-oncogene serine/threonine protein kinase | CENTRAL Signaling (JAK/STAT, TNF, VEGF, Insulin, Apelin, cAMP, mTOR etc.) Cancer (colorectal, pancreatic, breast, glioma, melanoma etc.) | |||
| hsa04371_Apelin_signaling_pathway | Infection (Hepatitis, Influenza, Tuberculosis, Salmonella etc.) has04210_Apoptosis, has04218_Cellular_senescence, has04510_Focal_adhesion, |
RAF1 |
SMAD4 | ||||
| hsa04390_Hippo_signaling_pathway | SMAD4 | ||||||
| hsa04550_Signaling_pathways_regulating_pluripotency_of_stem_cells | RAF1 | ID3 | SMAD4 | ||||
| hsa04625_C-type_lectin_receptor_signaling_pathway | RAF1 | ||||||
| hsa04630_JAK-STAT_signaling_pathway | RAF1 | STAT5B | |||||
| hsa04659_Th17_cell_differentiation | SMAD4# | Cdc42-interacting protein 4 | has04910_Insulin_signaling_pathway | ||||
| TTC14 * | Tetratricopeptide repeat protein 14 | - | |||||
| UBAP1 # | Ubiquitin-associated protein 1 | - | |||||
| has04540_Gap_junction, | UBXN8 * | UBX domain-containing protein 8 | has04141_Protein_processing_in_endoplasmic_reticulum | ||||
| ZNF334 * | Zinc finger protein 334 | has05168_Herpes_simplex_virus_1_infection | |||||
| ZNF639 # | Zinc finger protein 636 | - |
| hsa04068_FoxO_signaling_pathway | ||||
| RAF1 | ||||
| SMAD4 | ||||
| hsa04071_Sphingolipid_signaling_pathway | RAF1 | |||
| hsa04072_Phospholipase_D_signaling_pathway | RAF1 | |||
| hsa04150_mTOR_signaling_pathway | RAF1 | |||
| hsa04151_PI3K-Akt_signaling_pathway | RAF1 | |||
| STAT5B | ||||
| hsa04660_T_cell_receptor_signaling_pathway | RAF1 | |||
| hsa04662_B_cell_receptor_signaling_pathway | RAF1 | |||
| hsa04664_Fc_epsilon_RI_signaling_pathway | RAF1 | |||
| hsa04722_Neurotrophin_signaling_pathway | RAF1 | |||
| hsa04910_Insulin_signaling_pathway | RAF1 | TRIP10 | ||
| hsa04912_GnRH_signaling_pathway | RAF1 | |||
| hsa04915_Estrogen_signaling_pathway | RAF1 | |||
| hsa04917_Prolactin_signaling_pathway | RAF1 | STAT5B | ||
| hsa04919_Thyroid_hormone_signaling_pathway | RAF1 | ATP1B3 | ||
| hsa04921_Oxytocin_signaling_pathway | RAF1 | |||
| hsa04926_Relaxin_signaling_pathway | RAF1 | |||
| hsa04933_AGE-RAGE_signaling_pathway_in_diabetic_complications | SMAD4 | STAT5B | ||
| hsa04935_Growth_hormone_synthesis,_secretion_and_action | RAF1 | STAT5B | ||
| hsa05120_Epithelial_cell_signaling_in_Helicobacter_pylori_infection | GIT1 | |||
| hsa05161_Hepatitis_B | RAF1 | SMAD4 | STAT5B | |
| hsa05200_Pathways_in_cancer | RAF1 | RALBP1 | SMAD4 | STAT5B |
| hsa05210_Colorectal_cancer | RAF1 | SMAD4 | ||
| hsa05212_Pancreatic_cancer | RAF1 | RALBP1 | SMAD4 | |
| hsa05213_Endometrial_cancer | RAF1 | |||
| hsa05215_Prostate_cancer | RAF1 | |||
| hsa05219_Bladder_cancer | RAF1 | |||
| hsa05220_Chronic_myeloid_leukemia | RAF1 | SMAD4 | STAT5B | |
| hsa05221_Acute_myeloid_leukemia | RAF1 | STAT5B | ||
| hsa05223_Non-small_cell_lung_cancer | RAF1 | STAT5B | ||
| hsa05224_Breast_cancer | RAF1 | |||
| hsa05226_Gastric_cancer | RAF1 | SMAD4 |
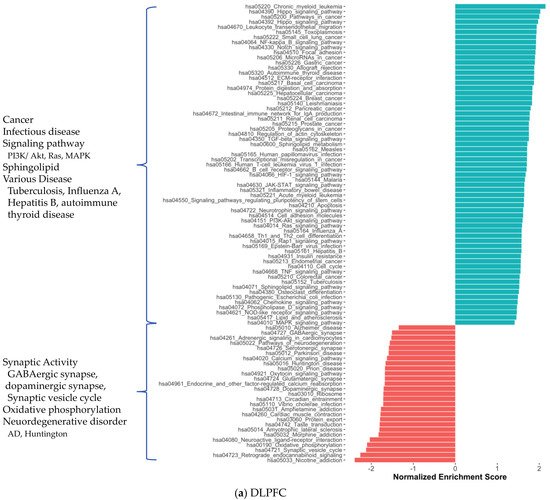
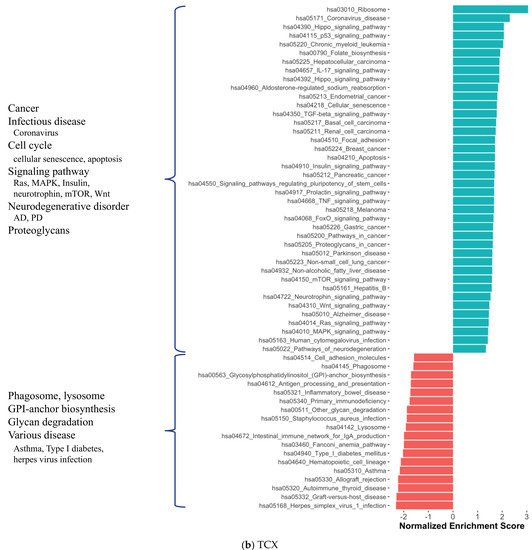
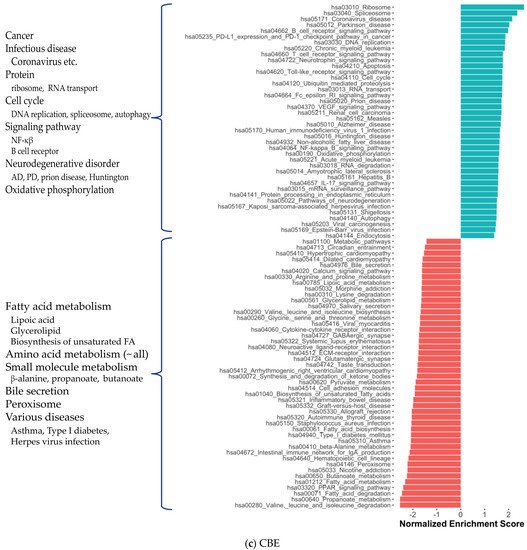
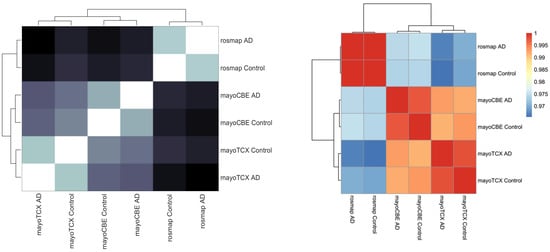
2. Co-Expression Network Analysis in Different Brain Regions of AD
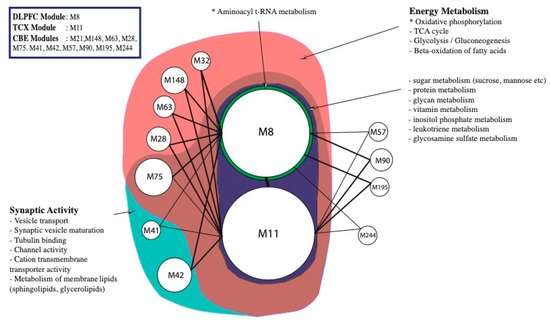
3. Alterations in Energy Metabolism, Chaperones and Synaptic Activity
| Tissues | Hub Genes (Top 10%) |
|---|---|
| DLPFC, TCX, CBE | AMIGO1, GPRASP2 |
| DLPFC, TCX | Cytoskeleton and its organisation ACTR3B, GABARAPL1, MARK1, NDEL1 Synaptic activity/plasticity AP2M1, ARHGEF9, CALM3, DLG3, GABRB3, L1CAM, NPTN Intra Golgi and retrograde Golgi-to-ER traffic |

| DLPFC-AD | DLPFC-Control | TCX-AD | TCX-Control | CBE-AD | CBE-Control | ||
|---|---|---|---|---|---|---|---|
| Number of Reactions | 5727 | 5773 | 5895 | 5845 | 5898 | 5826 | AP3M2, ARF3, CFAP36, KIFAP3, KLC1, NAPB, NSF, RAB6B Ubiquitin/Proteasome System DNAJC5, HSPA12A Glucose Metabolism/Oxidative phosphorylation ATP6V1A, ATP6V1B2, HK1, SEH1L, SLC25A14, SLC25A4, SLC9A6 Other ATL1, B4GAT1, BTRC, C1orf216, CDK14, CDK5R1CHN1, CISD1, CLSTN3, CNTNAP1, EID2, FAM234B, FAM49A, GLS, GOT1, GPI, GUCY1B3, INPP4A, JAZF1, MAGEE1, MAPK9, MLTT11, MOAP1, MYCBP2, NDFIBP1, NDRG3, NELL2, NMNAT2, OPCML, PCMT1, PFN2, PHACTR1, PNMA2, PNMAL1, PPPIR7, PPP3CB, PPP3R1, PREP, PREPL, PRKCE, RBFOX2, REEP1, REEP5, RTN3, SEPT6, SMAP2, SNAP91, SV2B, SYT13, TMEM246, TSPYL1, UBE2O, UBFD1, VDAC1, VDAC3, WDR7, YWHAG, YWHAZ |
| TCX, CBE | Cytoskeleton and its organisation PAK1 |
||||||
| DLPFC, CBE | Glucose Metabolism/Oxidative phosphorylation ATP6V1E1 Cytoskeleton and its organisation CDC42, DCTN2 Other ERLEC1, MPP1 |
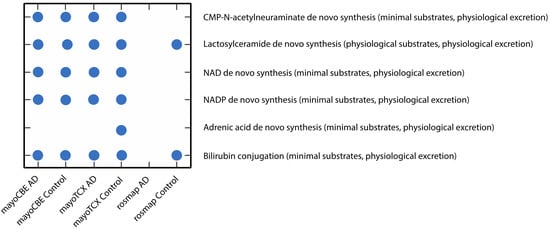
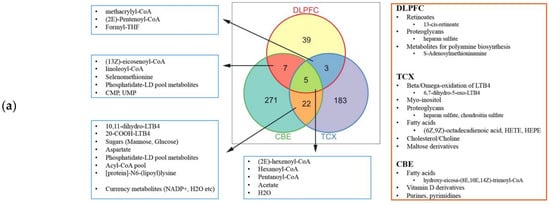
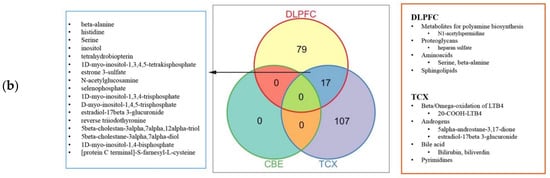
2.4. Metabolic Alterations in Different Brain Regions of AD
| Number of Metabolites | ||||||
| 4529 | 4593 | 4588 | 4541 | 4603 | 4542 | |
| Number of Genes | 2494 | 2516 | 2632 | 2615 | 2585 | 2592 |
References
- Levine, A.J.; Miller, J.A.; Shapshak, P.; Gelman, B.; Singer, E.J.; Hinkin, C.H.; Commins, D.; Morgello, S.; Grant, I.; Horvath, S. Systems analysis of human brain gene expression: Mechanisms for HIV-associated neurocognitive impairment and common pathways with Alzheimer’s disease. BMC Med. Genom. 2013, 6, 4.
- Borjabad, A.; Volsky, D.J. Common transcriptional signatures in brain tissue from patients with HIV-associated neurocognitive disorders, Alzheimer’s disease, and Multiple Sclerosis. J. Neuroimmune Pharmacol. 2012, 7, 914–926.
- Mastroeni, D.; Nolz, J.; Sekar, S.; Delvaux, E.; Serrano, G.; Cuyugan, L.; Liang, W.S.; Beach, T.G.; Rogers, J.; Coleman, P.D. Laser-captured microglia in the Alzheimer’s and Parkinson’s brain reveal unique regional expression profiles and suggest a potential role for hepatitis B in the Alzheimer’s brain. Neurobiol. Aging 2018, 63, 12–21.
- Cardoso, S.M.; Oliveira, C.R. Inhibition of NF-kB renders cells more vulnerable to apoptosis induced by amyloid beta peptides. Free Radic. Res. 2003, 37, 967–973.
- Zhao, Y.; Bhattacharjee, S.; Jones, B.M.; Hill, J.; Dua, P.; Lukiw, W.J. Regulation of neurotropic signaling by the inducible, NF-kB-sensitive miRNA-125b in Alzheimer’s disease (AD) and in primary human neuronal-glial (HNG) cells. Mol. Neurobiol. 2014, 50, 97–106.
- Religa, P.; Cao, R.; Religa, D.; Xue, Y.; Bogdanovic, N.; Westaway, D.; Marti, H.H.; Winblad, B.; Cao, Y. VEGF significantly restores impaired memory behavior in Alzheimer’s mice by improvement of vascular survival. Sci. Rep. 2013, 3, 2053.
- Gambuzza, M.E.; Sofo, V.; Salmeri, F.M.; Soraci, L.; Marino, S.; Bramanti, P. Toll-like receptors in Alzheimer’s disease: A therapeutic perspective. CNS Neurol. Disord. Drug Targets 2014, 13, 1542–1558.
- Kou, J.; Kovacs, G.G.; Höftberger, R.; Kulik, W.; Brodde, A.; Forss-Petter, S.; Hönigschnabl, S.; Gleiss, A.; Brügger, B.; Wanders, R.; et al. Peroxisomal alterations in Alzheimer’s disease. Acta Neuropathol. 2011, 122, 271–283.
- Jiang, Q.; Heneka, M.; Landreth, G.E. The role of peroxisome proliferator-activated receptor-gamma (PPARgamma) in Alzheimer’s disease: Therapeutic implications. CNS Drugs 2008, 22, 1–14.
- Gawel, K.; Labuz, K.; Jenda, M.; Silberring, J.; Kotlinska, J.H. Influence of cholinesterase inhibitors, donepezil and rivastigmine on the acquisition, expression, and reinstatement of morphine-induced conditioned place preference in rats. Behav. Brain Res. 2014, 268, 169–176.
- Klionsky, D.J.; Abdelmohsen, K.; Abe, A.; Abedin, M.J.; Abeliovich, H.; Arozena, A.A.; Adachi, H.; Adams, C.M.; Adams, P.D.; Adeli, K.; et al. Guidelines for the use and interpretation of assays for monitoring autophagy (3rd edition). Autophagy 2016, 12, 1–222.
- Feigin, V.L.; Nichols, E.; Alam, T.; Bannick, M.S.; Beghi, E.; Blake, N.; Culpepper, W.J.; Dorsey, E.R.; Elbaz, A.; Ellenbogen, R.G.; et al. Global, regional, and national burden of neurological disorders, 1990–2016: A systematic analysis for the Global Burden of Disease Study 2016. Lancet Neurol. 2019, 18, 459–480.
- Manavalan, A.; Mishra, M.; Sze, S.K.; Heese, K. Brain-site-specific proteome changes induced by neuronal P60TRP expression. Neurosignals 2013, 21, 129–149.
- Dagley, L.F.; Croft, N.P.; Isserlin, R.; Olsen, J.B.; Fong, V.; Emili, A.; Purcell, A.W. Discovery of novel disease-specific and membrane-associated candidate markers in a mouse model of multiple sclerosis. Mol. Cell. Proteom. 2014, 13, 679–700.
- Lee, K.W.; Bogenhagen, D.F. Assignment of 2’-O-methyltransferases to modification sites on the mammalian mitochondrial large subunit 16 S ribosomal RNA (rRNA). J. Biol. Chem. 2014, 289, 24936–24942.
- Lu, J.P.; Wang, Y.; Sliter, D.A.; Pearce, M.M.P.; Wojcikiewicz, R.J.H. RNF170 Protein, an Endoplasmic Reticulum Membrane Ubiquitin Ligase, Mediates Inositol 1,4,5-Trisphosphate Receptor Ubiquitination and Degradation *. J. Biol. Chem. 2011, 286, 24426–24433.
- Striessnig, J.; Pinggera, A.; Kaur, G.; Bock, G.; Tuluc, P. L-type Ca2+ channels in heart and brain. Wiley Interdiscip. Rev. Membr. Transp. Signal. 2014, 3, 15–38.
- Wren, L.M.; Jiménez-Jáimez, J.; Al-Ghamdi, S.; Al-Aama, J.Y.; Bdeir, A.; Al-Hassnan, Z.N.; Kuan, J.L.; Foo, R.Y.; Potet, F.; Johnson, C.N.; et al. Genetic Mosaicism in Calmodulinopathy. Circ. Genom. Precis. Med. 2019, 12, 375–385.
- Sosanya, N.M.; Cacheaux, L.P.; Workman, E.R.; Niere, F.; Perrone-Bizzozero, N.I.; Raab-Graham, K.F. Mammalian Target of Rapamycin (mTOR) Tagging Promotes Dendritic Branch Variability through the Capture of Ca2+/Calmodulin-dependent Protein Kinase II α (CaMKIIα) mRNAs by the RNA-binding Protein HuD. J. Biol. Chem. 2015, 290, 16357–16371.
- Paoletti, P.; Bellone, C.; Zhou, Q. NMDA receptor subunit diversity: Impact on receptor properties, synaptic plasticity and disease. Nat. Rev. Neurosci. 2013, 14, 383–400.
- Pamplona, R.; Borras, C.; Jové, M.; Pradas, I.; Ferrer, I.; Viña, J. Redox lipidomics to better understand brain aging and function. Free Radic. Biol. Med. 2019, 144, 310–321.
- Mattson, M.P.; Moehl, K.; Ghena, N.; Schmaedick, M.; Cheng, A. Intermittent metabolic switching, neuroplasticity and brain health. Nat. Rev. Neurosci. 2018, 19, 81–94.
- Papuć, E.; Rejdak, K. The role of myelin damage in Alzheimer’s disease pathology. Arch. Med. Sci. 2018, 16, 345–351.
- Alessenko, A.V.; Albi, E. Exploring Sphingolipid Implications in Neurodegeneration. Front. Neurol. 2020, 11, 437.
