Cardiovascular diseases (CVDs) are the leading cause of death worldwide. It is estimated that approximately 18.5 million people die annually on account of these diseases, with a third of these people dying under the age of 70 years. Identifying those most affected by CVDs and ensuring they receive the appropriate treatment can prevent premature deaths. Furthermore, the development of new therapeutic strategies and biomarkers with the potential to predict the progression of CVDs is fundamental to reducing mortality worldwide. CVDs can be defined as disorders that affect the heart or blood vessels such as heart failure, coronary heart disease, cerebrovascular disease, peripheral arterial disease, and congenital heart disease.
- aerobic training
- lncRNAs
- cardiovascular disease
- biomarkers
1. Introduction
2. LncRNAs in Cardiovascular Diseases
| lncRNA | CVDs | Association | References | |
|---|---|---|---|---|
| aHIF | MI | Regulation of the angiogenesis process and a biomarker. | [103] | [40] |
| aHIF | CHD | Biomarker. | [104] | [41] |
| AK098656 | AH | Regulation of arteries of resistance and a biomarker. | [105] | [42] |
| ANRIL | CHD | Susceptibility conferred by SNPs in the ANRIL locus on chromosome 9p | [106] | [43] |
| ANRIL | AH | Increase of susceptibility to higher systolic blood pressure conferred by polymorphisms. | [107] | [44] |
| ANRIL | MI | Protection of cardiomyocytes from hypoxia by acting on the miRNA-7-5p/SIRT1 axis; and biomarker to LV dysfunction. | [108,109,110] | [45][46][47] |
| ANRIL | HF | Biomarker. | [111] | [48] |
| APF | MI | Promotion of cardiomyocytes autophagy acting as a sponge for miRNA-188-3p. | [112] | [49] |
| APOA1-AS | CHD | Biomarker. | [104] | [41] |
| AWPPH | CHD | Biomarker. | [113] | [50] |
| BACE1-AS | HF | Promotion of ECs apoptosis. | [114] | [51] |
| BANCR | CHD | Promotion of VSMCs proliferation and migration. | [115] | [52] |
| CARL | MI | Reduction of mitochondrial fission and apoptosis acting as a sponge for miRNA-539. | [116] | [53] |
| CDR1AS | MI | Biomarker. | [117] | [54] |
| Chaer | HF | Induction of Pathological cardiac remodeling. | [118] | [55] |
| Chast | HF | Induction of Pathological cardiac remodeling. | [119] | [56] |
| CHRF | HF | Endogenous sponge to miRNA-489 activity. | [120] | [57] |
| CHROME | CHD | Regulation of cellular cholesterol homeostasis. | [121] | [58] |
| CoroMarker | CHD | Biomarker. | [122] | [59] |
| EGOT | HF | Biomarker. | [111] | [48] |
| FTX | MI | Regulation of cardiomyocytes apoptosis acting as a sponge for miRNA-29b-1-5. | [123] | [60] |
| GAS5 | AH | Regulation of ECs and VSMCs function acting as endogenous RNA competing of miRNA-21; and a biomarker. | [124,125] | [61][62] |
| GAS5 | MI | Protection of cardiomyocytes against hypoxic injury acting as a sponge for miRNA-142; promotion of the development and progression of the disease acting on the miRNA-525/CALM2 axis; and improves apoptosis by negatively regulating sema3a. | [126,127,128] | [63][64][65] |
| Giver | AH | Promotion of VSMCs dysfunction. | [129] | [66] |
| H19 | MI | Induction of cardiac remodeling; autophagy; and biomarker. | [130,131,132] | [67][68][69] |
| H19 | CHD | Biomarker. | [133,134] | [70][71] |
| H19 | HF | Regulation of cardiac hypertrophy; and a biomarker. | [111,135] | [48][72] |
| HEAT2 | HF | Biomarker. | [136] | [73] |
| HOTAIR | MI | Induction of cardioprotective acting as a sponge for miRNA-1 and as a biomarker. | [137] | [74] |
| HOTAIR | HF | Biomarker. | [111] | [48] |
| HOTTIP | CHD | Promotes ECs proliferation and migration. | [138] | [75] |
| HRCR | HF | Inhibition of cardiac hypertrophy acting as a sponge for miRNA-223. | [139] | [76] |
| KCNQ1OT1 | MI | Biomarker for left ventricular dysfunction. | [108] | [45] |
| LIPCAR | MI | Biomarker for cardiac remodeling. | [140] | [77] |
| LIPCAR | CHD | Biomarker. | [141] | [78] |
| LIPCAR | HF | Biomarker. | [140] | [77] |
| lincRNA-p21 | CHD | Regulation of cardiomyocytes apoptosis and proliferation. | [83,142] | [79][80] |
| LINC00968 | CHD | Promotion of ECs proliferation and migration acting as a sponge for miRNA-9. | [143] | [81] |
| lincRNA-ROR | HF | Regulation of cardiac hypertrophy acting as a sponge for miRNA-133. | [144] | [82] |
| Lnc-Ang362 | AH | Regulation of VSMCs proliferation through miRNA-221 and -222. | [145] | [83] |
| Lnc-Ang362 | MI | Promotion of cardiac fibrosis. | [146] | [84] |
| LOC285194 | HF | Biomarker. | [111] | [48] |
| MALAT1 | MI | Regulation of cardiomyocytes apoptosis and autophagy through miRNA-558; and biomarker. | [132,147,148] | [69][85][86] |
| MALAT1 | CHD | Biomarker. | [149] | [87] |
| MDRL | MI | Reduction of mitochondrial fission and apoptosis acting as a sponge for miRNA-361. | [150] | [88] |
| MEG3 | MI | Regulation of cardiomyocytes apoptosis. | [151] | [89] |
| MEG3 | HF | Regulation of cardiac fibrosis and diastolic dysfunction. | [152] | [90] |
| MHRT | MI | Regulation of cardiomyocytes apoptosis; and biomarker. | [153] | [91] |
| MHRT | HF | Regulation of chromatin remodelers; and biomarker. | [154,155] | [92][93] |
| MIAT | MI | Regulation of cardiac hypertrophy and fibrosis acting as a sponge for miRNA-150 and -93. | [102,156,157] | [39][94][95] |
| MIAT | CHD | Biomarker. | [149] | [87] |
| MIAT | HF | Regulation of cardiac hypertrophy acting as a sponge for miRNA-150. | [157] | [95] |
| Mirt1/2 | MI | Regulation of cardiac remodeling. | [158] | [96] |
| n379519 | MI | Promotion of cardiac fibrosis through miRNA-30. | [159] | [97] |
| NEXN-AS1 | CHD | Mitigation of atherosclerosis. | [160] | [98] |
| NONRATT021972 | MI | Promotion of cardiac function. | [161] | [99] |
| NR_027032 | AH | Biomarker. | [162] | [100] |
| NR_034083 | AH | Biomarker. | [162] | [100] |
| NR_104181 | AH | Biomarker. | [162] | [100] |
| NRF | MI | Regulation of cardiomyocytes necrosis. | [163] | [101] |
| NRON | HF | Biomarker. | [155] | [93] |
| PCFL | MI | Promotion of cardiac fibrosis through miRNA-378. | [164] | [102] |
| RMRP | HF | Biomarker. | [111] | [48] |
| RNY5 | HF | Biomarker. | [111] | [48] |
| SMILR | CHD | Biomarker. | [165] | [103] |
| SOX2-OT | HF | Biomarker. | [111] | [48] |
| SRA1 | HF | Biomarker. | [111] | [48] |
| TTTY15 | MI | Induction of cardiomyocyte injury by hypoxia targeting miRNA-455. | [166] | [104] |
| UCA1 | MI | Biomarker. | [167,168] | [105][106] |
| UIHTC | MI | Promotion of mitochondrial function. | [169] | [107] |
| Wisper | MI | Regulation of cardiac fibroblast. | [170] | [108] |
| ZFAS1 | MI | Induction of cardiomyocyte apoptosis; cardiac contractility reduction; and biomarker. | [117,169,171] | [54][107][109] |
Given the specificity of expression of lncRNAs, it would be careless to think that the dysregulation of the expression of these molecules in cardiac pathological processes, even if the molecular mechanism behind them is not exactly understood, was a mere coincidence [69][110]. The poor conservation of these interspecies transcripts, however, makes it difficult to translate findings in rodent models for human applications; however, several studies have shown promising results regarding the prognosis of CVDs and new therapies from the modulation of cardiac lncRNAs [33,34,70,89,90,172,173][32][33][111][112][113][114][115]. We summarize the lncRNAs and the CVDs (Figure 21).
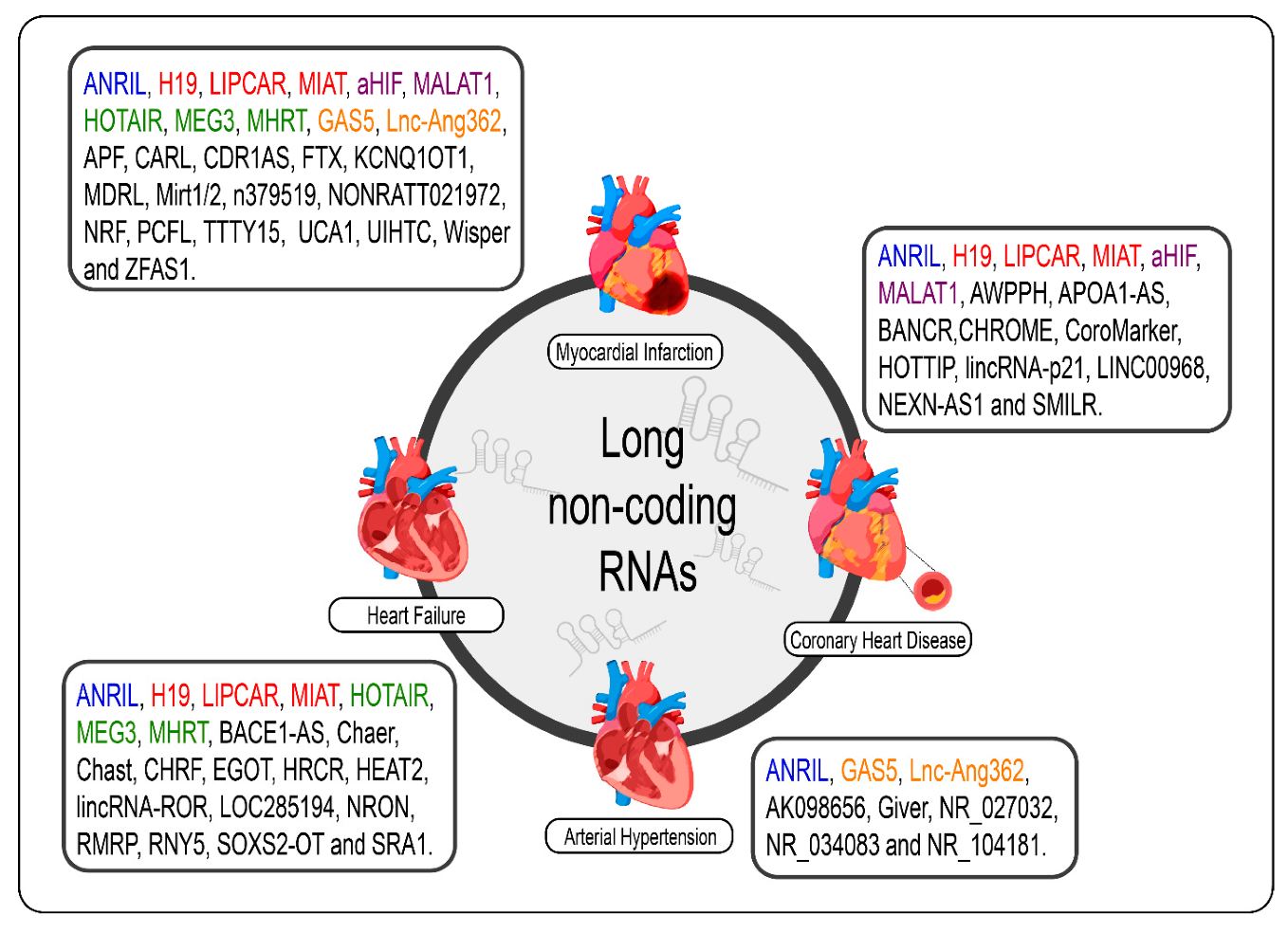
Figure 21. lncRNAs are differentially expressed in cardiovascular diseases such as heart failure, myocardial infarction, coronary artery disease, and arterial hypertension. The lncRNAs marked in blue are the same present in myocardial infarction, coronary artery disease, heart failure, and arterial hypertension; those marked in red are the same present in myocardial infarction, coronary artery disease, and heart failure; those marked in green are the same present in myocardial infarction and heart failure, those marked in orange are present in myocardial infarction and arterial hypertension; and the purple present in myocardial infarction and coronary artery disease.
3. LncRNAs in Cardiovascular Diseases: Challenges and Future Perspectives
4. Conclusions
References
- Sharma, S.; Merghani, A.; Mont, L. Exercise and the Heart: The Good, the Bad, and the Ugly. Eur. Hear. J. 2015, 36, 1445–1453.
- Pedersen, B.K.; Saltin, B. Exercise as Medicine-Evidence for Prescribing Exercise as Therapy in 26 Different Chronic Diseases. Scand. J. Med. Sci. Sports 2015, 25, 1–72.
- Dunstan, D.W.; Dogra, S.; Carter, S.E.; Owen, N. Sit Less and Move More for Cardiovascular Health: Emerging Insights and Opportunities. Nat. Rev. Cardiol. 2021, 18, 637–648.
- ENCODE: Encyclopedia of DNA Elements. Available online: https://www.encodeproject.org/ (accessed on 28 August 2021).
- Reuter, J.A.; Spacek, D.V.; Snyder, M.P. High-Throughput Sequencing Technologies. Mol. Cell 2015, 58, 586–597.
- Delihas, N. Discovery and Characterization of the First Non-Coding RNA That Regulates Gene Expression, micFRNA: A Historical Perspective. World J. Biol. Chem. 2015, 6, 272–280.
- Ludwig, M. Non-Coding DNA Evolution: Junk DNA Revisited. Encycl. Evol. Biol. 2016, 6, 124–129.
- Palazzo, A.F.; Lee, E.S. Non-Coding RNA: What Is Functional and What Is Junk? Front. Genet. 2015, 6, 2.
- Ponjavic, J.; Ponting, C.P.; Lunter, G. Functionality or Transcriptional Noise? Evidence for Selection within Long Noncoding RNAs. Genome Res. 2007, 17, 556–565.
- Mattick, J.; Makunin, I.V. Non-Coding RNA. Hum. Mol. Genet. 2006, 15, 17–29.
- Churchman, L.S. Not Just Noise: Genomics and Genetics Bring Long Noncoding RNAs into Focus. Mol. Cell 2017, 65, 1–2.
- Xue, Y.; Chen, R.; Qu, L.; Cao, X. Noncoding RNA: From Dark Matter to Bright Star. Sci. China Life Sci. 2020, 63, 463–468.
- Mongelli, A.; Martelli, F.; Farsetti, A.; Gaetano, C. The Dark That Matters: Long Non-Coding RNAs as Master Regulators of Cellular Metabolism in Non-communicable Diseases. Front. Physiol. 2019, 10, 369.
- Kaikkonen, M.U.; Lam, M.T.; Glass, C.K. Non-Coding RNAs as Regulators of Gene Expression and Epigenetics. Cardiovasc. Res. 2011, 90, 430–440.
- Wei, J.-W.; Huang, K.; Yang, C.; Kang, C.-S. Non-Coding RNAs as Regulators in Epigenetics. Oncol. Rep. 2016, 37, 3–9.
- Chen, Y.-C.A.; Aravin, A.A. Non-Coding RNAs in Transcriptional Regulation. Curr. Mol. Biol. Rep. 2015, 1, 10–18.
- Patil, V.S.; Zhou, R.; Rana, T.M. Gene Regulation by Non-Coding RNAs. Crit. Rev. Biochem. Mol. Biol. 2013, 49, 16–32.
- O’Brien, J.; Hayder, H.; Zayed, Y.; Peng, C. Overview of MicroRNA Biogenesis, Mechanisms of Actions, and Circulation. Front. Endocrinol. 2018, 9, 402.
- Olson, E.N. MicroRNAs as Therapeutic Targets and Biomarkers of Cardiovascular Disease. Sci. Transl. Med. 2014, 6, 239ps3.
- Huang, W. MicroRNAs: Biomarkers, Diagnostics, and Therapeutics. Methods Mol. Biol. 2017, 1617, 57–67.
- Gomes, C.P.; de Gonzalo-Calvo, D.; Toro, R.; Fernandes, T.; Theisen, D.; Wang, D.-Z.; Devaux, Y. Non-Coding RNAs and Exercise: Pathophysiological Role and Clinical Application in the Cardiovascular System. Clin. Sci. 2018, 132, 925–942.
- Da Silva, G.J.; Bye, A.; el Azzouzi, H.; Wisløff, U. MicroRNAs as Important Regulators of Exercise Adaptation. Prog. Cardiovasc. Dis. 2017, 60, 130–151.
- Altana, V.; Geretto, M.; Pulliero, A. MicroRNAs and Physical Activity. MicroRNA 2015, 4, 74–85.
- Meurer, S.; Krüger, K.; Mooren, F. MicroRNAs unter Einfluss Körperlicher Belastung. Ger. J. Sports Med. 2016, 2016, 27–34.
- Yao, R.; Wang, Y.; Chen, L.-L. Cellular Functions of Long Noncoding RNAs. Nat. Cell Biol. 2019, 21, 542–551.
- Kapusta, A.; Feschotte, C. Volatile Evolution of Long Noncoding RNA Repertoires: Mechanisms and Biological Implications. Trends Genet. 2014, 30, 439–452.
- Statello, L.; Guo, C.-J.; Chen, L.-L.; Huarte, M. Gene Regulation by Long Non-Coding RNAs and Its Biological Functions. Nat. Rev. Mol. Cell Biol. 2020, 22, 96–118.
- Sun, M.; Kraus, W.L. From Discovery to Function: The Expanding Roles of Long Non-Coding RNAs in Physiology and Disease. Endocr. Rev. 2015, 36, 25–64.
- Dhanoa, J.K.; Sethi, R.S.; Verma, R.; Arora, J.S.; Mukhopadhyay, C.S. Long Non-Coding RNA: Its Evolutionary Relics and Biological Implications in Mammals: A Review. J. Anim. Sci. Technol. 2018, 60, 1–10.
- Espinosa, J.M. On the Origin of lncRNAs: Missing Link Found. Trends Genet. 2017, 33, 660–662.
- Jarroux, J.; Morillon, A.; Pinskaya, M. History, Discovery, and Classification of lncRNAs. Adv. Exp. Med. Biol. 2017, 1008, 1–46.
- McMullen, J.R.; Drew, B.G. Long Non-Coding RNAs (lncRNAs) in Skeletal and Cardiac Muscle: Potential Therapeutic and Diagnostic Targets? Clin. Sci. 2016, 130, 2245–2256.
- Gomes, C.P.C.; Spencer, H.; Ford, K.L.; Michel, L.Y.M.; Baker, A.H.; Emanueli, C.; Balligand, J.-L.; Devaux, Y. The Function and Therapeutic Potential of Long Non-Coding RNAs in Cardiovascular Development and Disease. Mol. Ther.-Nucleic Acids 2017, 8, 494–507.
- Bonilauri, B.; Dallagiovanna, B. Long Non-Coding RNAs Are Differentially Expressed after Different Exercise Training Programs. Front. Physiol. 2020, 11.
- DiStefano, J.K. The Emerging Role of Long Noncoding RNAs in Human Disease. Methods Mol. Biol. 2018, 1706, 91–110.
- Ismail, N.; Abdullah, N.; Murad, N.A.; Jamal, R.; Sulaiman, S. Long Non-Coding RNAs (lncRNAs) in Cardiovascular Disease Complication of Type 2 Diabetes. Diagnostics 2021, 11, 145.
- Schmitt, A.M.; Chang, H.Y. Long Noncoding RNAs in Cancer Pathways. Cancer Cell 2016, 29, 452–463.
- Liu, S.J.; Dang, H.X.; Lim, D.A.; Feng, F.Y.; Maher, C.A. Long Noncoding RNAs in Cancer Metastasis. Nat. Rev. Cancer 2021, 21, 446–460.
- Ishii, N.; Ozaki, K.; Sato, H.; Mizuno, H.; Saito, S.; Takahashi, A.; Miyamoto, Y.; Ikegawa, S.; Kamatani, N.; Hori, M.; et al. Identification of a Novel Non-Coding RNA, MIAT, That Confers Risk of Myocardial Infarction. J. Hum. Genet. 2006, 51, 1087–1099.
- Semenza, G.L. Hypoxia-Inducible Factor 1 and Cardiovascular Disease. Annu. Rev. Physiol. 2014, 76, 39–56.
- Zhang, Y.; Zhang, L.; Wang, Y.; Ding, H.; Xue, S.; Yu, H.; Hu, L.; Qi, H.; Wang, Y.; Zhu, W.; et al. KCNQ 1 OT 1, HIF 1A-AS 2 and APOA 1-AS Are Promising Novel Biomarkers for Diagnosis of Coronary Artery Disease. Clin. Exp. Pharmacol. Physiol. 2019, 46, 635–642.
- Jin, L.; Lin, X.; Yang, L.; Fan, X.; Wang, W.; Li, S.; Li, J.; Liu, X.; Bao, M.; Cui, X.; et al. AK098656, a Novel Vascular Smooth Muscle Cell–Dominant Long Noncoding RNA, Promotes Hypertension. Hypertension 2018, 71, 262–272.
- Broadbent, H.M.; Peden, J.F.; Lorkowski, S.; Goel, A.; Ongen, H.; Green, F.; Clarke, R.; Collins, R.; Franzosi, M.G.; Tognoni, G.; et al. Susceptibility to Coronary Artery Disease and Diabetes Is Encoded by Distinct, Tightly Linked SNPs in the ANRIL Locus on Chromosome 9p. Hum. Mol. Genet. 2007, 17, 806–814.
- Bayoglu, B.; Yuksel, H.; Cakmak, H.A.; Dirican, A.; Cengiz, M. Polymorphisms in the Long Non-Coding RNA CDKN2B-AS1 May Contribute to Higher Systolic Blood Pressure Levels in Hyper-Tensive Patients. Clin. Biochem. 2016, 49, 821–827.
- Vausort, M.; Wagner, D.R.; Devaux, Y. Long Noncoding RNAs in Patients with Acute Myocardial Infarction. Circ. Res. 2014, 115, 668–677.
- Ahmed, W.; Ali, I.S.; Riaz, M.; Younas, A.; Sadeque, A.; Niazi, A.K.; Niazi, S.H.; Ali, S.H.B.; Azam, M.; Qamar, R. Association of ANRIL Polymorphism (rs1333049:C>G) with Myocardial Infarction and Its Pharmacogenomic Role in Hypercholesterolemia. Gene 2013, 515, 416–420.
- Shu, L.; Zhang, W.; Huang, C.; Huang, G.; Su, G.; Xu, J. lncRNA ANRIL protects H9c2 cells against hypoxia-induced injury through targeting the miR-7-5p/SIRT1 axis. J. Cell. Physiol. 2019, 235, 1175–1183.
- Greco, S.; Zaccagnini, G.; Perfetti, A.; Fuschi, P.; Valaperta, R.; Voellenkle, C.; Castelvecchio, S.; Gaetano, C.; Finato, N.; Beltrami, A.P.; et al. Long Noncoding Rna Dysregulation in Ischemic Heart Failure. J. Transl. Med. 2016, 14, 1–14.
- Wang, K.; Liu, C.-Y.; Zhou, L.-Y.; Wang, J.; Wang, M.; Zhao, B.; Zhao, W.-K.; Jian-Xun, W.; Yan-Fang, Z.; Zhang, X.-J.; et al. APF lncRNA Regulates Autophagy and Myocardial Infarction by Targeting miR-188-3p. Nat. Commun. 2015, 6, 6779.
- Tang, T.-T.; Wang, B.-Q. Clinical Significance of lncRNA-AWPPH in Coronary Artery Diseases. Eur. Rev. Med. Pharmacol. Sci. 2020, 24, 11747–11751.
- Greco, S.; Zaccagnini, G.; Fuschi, P.; Voellenkle, C.; Carrara, M.; Sadeghi, I.; Bearzi, C.; Maimone, B.; Castelvecchio, S.; Stellos, K.; et al. Increased BACE1-AS Long Noncoding RNA and β-Amyloid Levels in Heart Failure. Cardiovasc. Res. 2017, 113, 453–463.
- Li, H.; Liu, X.; Zhang, L.; Li, X. LncRNA BANCR Facilitates Vascular Smooth Muscle Cell Proliferation and Migration through JNK Pathway. Oncotarget 2017, 8, 114568–114575.
- Wang, K.; Long, B.; Zhou, L.-Y.; Liu, F.; Zhou, Q.-Y.; Liu, C.-Y.; Fan, Y.-Y.; Li, P.-F. CARL lncRNA Inhibits Anoxia-Induced Mitochondrial Fission and Apoptosis in Cardiomyocytes by Impairing miR-539-Dependent PHB2 Downregulation. Nat. Commun. 2014, 5, 3596.
- Zhang, Y.; Sun, L.; Xuan, L.; Pan, Z.; Li, K.; Liu, S.; Huang, Y.; Zhao, X.; Huang, L.; Wang, Z.; et al. Reciprocal Changes of Circulating Long Non-Coding RNAs ZFAS1 and CDR1AS Predict Acute Myocardial Infarction. Sci. Rep. 2016, 6, 22384.
- Wang, Z.; Zhang, X.-J.; Ji, Y.-X.; Zhang, P.; Deng, K.-Q.; Gong, J.; Ren, S.; Wang, X.; Chen, I.; Wang, H.; et al. The Long Noncoding RNA CHAER Defines an Epigenetic Checkpoint in Cardiac Hypertrophy. Nat. Med. 2016, 22, 1131–1139.
- Viereck, J.; Kumarswamy, R.; Foinquinos, A.; Xiao, K.; Avramopoulos, P.; Kunz, M.; Dittrich, M.; Maetzig, T.; Zimmer, K.; Remke, J.; et al. Long Noncoding RNA CHAST Promotes Cardiac Remodeling. Sci. Transl. Med. 2016, 8, 326ra22.
- Wang, K.; Liu, F.; Zhou, L.-Y.; Long, B.; Yuan, S.-M.; Wang, Y.; Liu, C.-Y.; Sun, T.; Zhang, X.-J.; Li, P.-F. The Long Noncoding RNA CHRF Regulates Cardiac Hypertrophy by Targeting miR-489. Circ. Res. 2014, 114, 1377–1388.
- Hennessy, E.J.; Van Solingen, C.; Scacalossi, K.R.; Ouimet, M.; Afonso, M.S.; Prins, J.; Koelwyn, G.J.; Sharma, M.; Ramkhelawon, B.; Carpenter, S.; et al. The Long Noncoding RNA CHROME Regulates Cholesterol Homeostasis in Primates. Nat. Metab. 2018, 1, 98–110.
- Yang, Y.; Cai, Y.; Wu, G.; Chen, X.; Liu, Y.; Wang, X.; Yu, J.; Li, C.; Chen, X.; Jose, P.A.; et al. Plasma Long Non-Coding RNA, CoroMarker, a Novel Biomarker for Diagnosis of Coronary Artery Disease. Clin. Sci. 2015, 129, 675–685.
- Long, B.; Li, N.; Xu, X.-X.; Li, X.-X.; Xu, X.-J.; Guo, D.; Zhang, D.; Wu, Z.-H.; Zhang, S.-Y. Long Noncoding RNA FTX Regulates Cardiomyocyte Apoptosis by Targeting miR-29b-1-5p and Bcl2l2. Biochem. Biophys. Res. Commun. 2018, 495, 312–318.
- Wang, Y.-N.; Shan, K.; Yao, M.-D.; Yao, J.; Wang, J.-J.; Li, X.; Liu, B.; Zhang, Y.-Y.; Ji, Y.; Jiang, Q.; et al. Long Noncoding RNA-GAS5: A Novel Regulator of Hypertension-Induced Vascular Remodeling. Hypertension 2016, 68, 736–748.
- Liu, K.; Liu, C.; Zhang, Z. lncRNA GAS5 Acts as a ceRNA for miR-21 in Suppressing PDGF-BB-Induced Proliferation and Migration in Vascular Smooth Muscle Cells. J. Cell. Biochem. 2019, 120, 15233–15240.
- Du, J.; Yang, S.-T.; Liu, J.; Zhang, K.-X.; Leng, J.-Y. Silence of LncRNA GAS5 Protects Cardiomyocytes H9c2 against Hypoxic Injury via Sponging miR-142-5p. Mol. Cells 2019, 42, 397–405.
- Zhang, Y.; Hou, Y.-M.; Gao, F.; Xiao, J.-W.; Li, C.-C.; Tang, Y. lncRNA GAS5 Regulates Myocardial Infarction by Targeting the miR-525-5p/CALM2 Axis. J. Cell. Biochem. 2019, 120, 18678–18688.
- Hao, S.; Liu, X.; Sui, X.; Pei, Y.; Liang, Z.; Zhou, N. Long Non-Coding RNA GAS5 Reduces Cardiomyocyte Apoptosis Induced by MI through sema3a. Int. J. Biol. Macromol. 2018, 120, 371–377.
- Das, S.; Zhang, E.; Senapati, P.; Amaram, V.; Reddy, M.A.; Stapleton, K.; Leung, A.; Lanting, L.; Wang, M.; Chen, Z.; et al. A Novel Angiotensin II–Induced Long Noncoding RNA Giver Regulates Oxidative Stress, Inflammation, and Proliferation in Vascular Smooth Muscle Cells. Circ. Res. 2018, 123, 1298–1312.
- Zhou, M.; Zou, Y.-G.; Xue, Y.-Z.; Wang, X.-H.; Gao, H.; Dong, H.-W.; Zhang, Q. Long Non-Coding RNA H19 Protects Acute Myocardial Infarction through Activating Autophagy in Mice. Eur. Rev. Med. Pharmacol. Sci. 2018, 22, 5647–5651.
- Choong, O.K.; Chen, C.-Y.; Zhang, J.; Lin, J.-H.; Lin, P.-J.; Ruan, S.-C.; Kamp, T.J.; Hsieh, P.C. Hypoxia-Induced H19/YB-1 Cascade Modulates Cardiac Remodeling after Infarction. Theranostics 2019, 9, 6550–6567.
- Wang, X.-M.; Li, X.-M.; Song, N.; Zhai, H.; Gao, X.-M.; Yang, Y.-N. Long Non-Coding RNAs H19, MALAT1 and MIAT as Potential Novel Biomarkers for Diagnosis of Acute Myocardial Infarction. Biomed. Pharmacother. 2019, 118, 109208.
- Yao, Y.; Xiong, G.; Jiang, X.; Song, T. The Overexpression of lncRNA H19 as a Diagnostic Marker for Coronary Artery Disease. Rev. Assoc. Med. Bras. 2019, 65, 110–117.
- Gao, W.; Zhu, M.; Wang, H.; Zhao, S.; Zhao, D.; Yang, Y.; Wang, Z.-M.; Wang, F.; Yang, Z.-J.; Lu, X.; et al. Association of Polymorphisms in Long Non-Coding RNA H19 with Coronary Artery Disease Risk in a Chinese Population. Mutat. Res. Mol. Mech. Mutagen. 2015, 772, 15–22.
- Liu, L.; An, X.; Li, Z.; Song, Y.; Li, L.; Zuo, S.; Liu, N.; Yang, G.; Wang, H.; Cheng, X.; et al. The H19 Long Noncoding RNA Is a Novel Negative Regulator of Cardiomyocyte Hypertrophy. Cardiovasc. Res. 2016, 111, 56–65.
- Boeckel, J.-N.; Perret, M.F.; Glaser, S.F.; Seeger, T.; Heumüller, A.W.; Chen, W.; John, D.; Kokot, K.E.; Katus, H.A.; Haas, J.; et al. Identification and Regulation of the Long Non-Coding RNA Heat2 in Heart Failure. J. Mol. Cell. Cardiol. 2019, 126, 13–22.
- Gao, L.; Liu, Y.; Guo, S.; Yao, R.; Wu, L.; Xiao, L.; Wang, Z.; Liu, Y.; Zhang, Y. Circulating Long Noncoding RNA HOTAIR is an Essential Mediator of Acute Myocardial Infarction. Cell. Physiol. Biochem. 2017, 44, 1497–1508.
- Liao, B.; Chen, R.; Lin, F.; Mai, A.; Chen, J.; Li, H.; Xu, Z.; Dong, S. Long Noncoding RNA HOTTIP Promotes Endothelial Cell Proliferation and Migration via Activation of the Wnt/β-Catenin Pathway. J. Cell. Biochem. 2017, 119, 2797–2805.
- Devaux, Y.; Creemers, E.E.; Boon, R.A.; Werfel, S.; Thum, T.; Engelhardt, S.; Dimmeler, S.; Squire, I. Circular RNAs in Heart Failure. Eur. J. Hear. Fail. 2017, 19, 701–709.
- Kumarswamy, R.; Bauters, C.; Volkmann, I.; Maury, F.; Fetisch, J.; Holzmann, A.; Lemesle, G.; de Groote, P.; Pinet, F.; Thum, T. Circulating Long Noncoding RNA, LIPCAR, Predicts Survival in Patients with Heart Failure. Circ. Res. 2014, 114, 1569–1575.
- Zhang, Z.; Gao, W.; Long, Q.-Q.; Zhang, J.; Lian-Sheng, W.; Liu, D.-C.; Yan, J.-J.; Yang, Z.-J.; Wang, L.-S. Increased Plasma Levels of lncRNA H19 and LIPCAR Are Associated with Increased Risk of Coronary Artery Disease in a Chinese Population. Sci. Rep. 2017, 7, 1–9.
- Huarte, M.; Guttman, M.; Feldser, D.; Garber, M.; Koziol, M.J.; Kenzelmann-Broz, D.; Khalil, A.M.; Zuk, O.; Amit, I.; Rabani, M.; et al. A Large Intergenic Noncoding RNA Induced by p53 Mediates Global Gene Repression in the p53 Response. Cell 2010, 142, 409–419.
- Tang, S.-S.; Cheng, J.; Cai, M.-Y.; Yang, X.-L.; Liu, X.G.; Zheng, B.-Y.; Xiong, X.-D. Association of lincRNA-p21Haplotype with Coronary Artery Disease in a Chinese Han Population. Dis. Markers 2016, 2016, 1–7.
- Wang, X.; Zhao, Z.; Zhang, W.; Wang, Y. Long Noncoding RNA LINC00968 Promotes Endothelial Cell Proliferation and Migration via Regulating miR-9-3p Expression. J. Cell. Biochem. 2019, 120, 8214–8221.
- Jiang, F.; Zhou, X.; Huang, J. Long Non-Coding RNA-ROR Mediates the Reprogramming in Cardiac Hypertrophy. PLoS ONE 2016, 11, e0152767.
- Leung, A.; Trac, C.; Jin, W.; Lanting, L.; Akbany, A.; Sætrom, P.; Schones, D.E.; Natarajan, R. Novel Long Noncoding RNAs Are Regulated by Angiotensin II in Vascular Smooth Muscle Cells. Circ. Res. 2013, 113, 266–278.
- Chen, G.; Huang, S.; Song, F.; Zhou, Y.; He, X. Lnc-Ang362 Is a Pro-Fibrotic Long Non-Coding RNA Promoting Cardiac Fibrosis after Myocardial Infarction by Suppressing Smad7. Arch. Biochem. Biophys. 2020, 685, 108354.
- Guo, X.; Wu, X.; Han, Y.; Tian, E.; Cheng, J. LncRNA MALAT1 Protects Cardiomyocytes from Isoproterenol-Induced Apoptosis through Sponging miR-558 to Enhance ULK1-Mediated Protective Autophagy. J. Cell. Physiol. 2018, 234, 10842–10854.
- Hu, L.; Xu, Y.-N.; Wang, Q.; Liu, M.-J.; Zhang, P.; Zhao, L.-T.; Liu, F.; Zhao, D.-Y.; Pei, H.-N.; Yao, X.-B.; et al. Aerobic Exercise Improves Cardiac Function in Rats with Chronic Heart Failure through Inhibition of the Long Non-Coding RNA Metastasis-Associated Lung Adenocarcinoma Transcript 1 (MALAT1). Ann. Transl. Med. 2021, 9, 340.
- Toraih, E.; El-Wazir, A.; Alghamdi, S.A.; Alhazmi, A.S.; El-Wazir, M.; Abdel-Daim, M.; Fawzy, M.S. Association of Long Non-Coding RNA MIAT and MALAT1 Expression Profiles in Peripheral Blood of Coronary Artery Disease Patients with Previous Cardiac Events. Genet. Mol. Biol. 2019, 42, 509–518.
- Wang, K.; Sun, T.; Li, N.; Wang, Y.; Wang, J.; Zhou, L.-Y.; Long, B.; Liu, C.-Y.; Liu, F.; Li, P.-F. MDRL lncRNA Regulates the Processing of miR-484 Primary Transcript by Targeting miR-361. PLoS Genet. 2014, 10, e1004467.
- Wu, H.; Zhao, Z.-A.; Liu, J.; Hao, K.; Yu, Y.; Han, X.; Li, J.; Wang, Y.; Lei, W.; Dong, N.; et al. Long Noncoding RNA Meg3 Regulates Cardiomyocyte Apoptosis in Myocardial Infarction. Gene Ther. 2018, 25, 511–523.
- Piccoli, M.-T.; Gupta, S.K.; Viereck, J.; Foinquinos, A.; Samolovac, S.; Kramer, F.L.; Garg, A.; Remke, J.; Zimmer, K.; Batkai, S.; et al. Inhibition of the Cardiac Fibroblast–Enriched lncRNA Meg3 Prevents Cardiac Fibrosis and Diastolic Dysfunction. Circ. Res. 2017, 121, 575–583.
- Zhang, J.; Gao, C.; Meng, M.; Tang, H. Long Noncoding RNA MHRT Protects Cardiomyocytes against H2O2-Induced Apoptosis. Biomol. Ther. 2016, 24, 19–24.
- Han, P.; Li, W.; Lin, C.-H.; Yang, J.; Shang, C.; Nuernberg, S.T.; Jin, K.K.; Xu, W.; Lin, C.-Y.; Lin, C.-J.; et al. A Long Noncoding Rna Protects the Heart from Pathological Hypertrophy. Nat. Cell Biol. 2014, 514, 102–106.
- Xuan, L.; Sun, L.; Zhang, Y.; Huang, Y.; Hou, Y.; Li, Q.; Guo, Y.; Feng, B.; Cui, L.; Wang, X.; et al. Circulating Long Non-Coding Rnas Nron and MHRT as Novel Predictive Biomarkers of Heart Failure. J. Cell. Mol. Med. 2017, 21, 1803–1814.
- Qu, X.; Du, Y.; Shu, Y.; Gao, M.; Sun, F.; Luo, S.; Yang, T.; Zhan, L.; Yuan, Y.; Chu, W.; et al. MIAT Is a Pro-Fibrotic Long Non-Coding RNA Governing Cardiac Fibrosis in Post-Infarct Myocardium. Sci. Rep. 2017, 7, srep42657.
- Zhu, X.-H.; Yuan, Y.-X.; Rao, S.-L.; Wang, P. LncRNA MIAT Enhances Cardiac Hypertrophy Partly through Sponging miR-150. Eur. Rev. Med. Pharmacol. Sci. 2016, 20, 3653.
- Zangrando, J.; Zhang, L.; Vausort, M.; Maskali, F.; Marie, P.-Y.; Wagner, D.R.; Devaux, Y. Identification of Candidate Long Non-Coding RNAs in Response to Myocardial Infarction. BMC Genom. 2014, 15, 460.
- Wang, X.; Yong, C.; Yu, K.; Yu, R.; Zhang, R.; Yu, L.; Li, S.; Cai, S. Long Noncoding RNA (lncRNA) n379519 Promotes Cardiac Fibrosis in Post-Infarct Myocardium by Targeting miR-30. Med. Sci. Monit. 2018, 24, 3958–3965.
- Hu, Y.-W.; Guo, F.-X.; Xu, Y.-J.; Li, P.; Lu, Z.-F.; McVey, D.G.; Zheng, L.; Wang, Q.; Ye, J.; Kang, C.-M.; et al. Long Noncoding RNA NEXN-AS1 Mitigates Atherosclerosis by Regulating the Actin-Binding Protein NEXN. J. Clin. Investig. 2019, 129, 1115–1128.
- Tu, G.; Zou, L.; Liu, S.; Wu, B.; Lv, Q.; Wang, S.; Xue, Y.; Zhang, C.; Yi, Z.; Zhang, X.; et al. Long Noncoding NONRATT021972 siRNA Normalized Abnormal Sympathetic Activity Mediated by the Upregulation of P2X7 Receptor in Superior Cervical Ganglia after Myocardial Ischemia. Purinergic Signal. 2016, 12, 521–535.
- Chen, S.; Chen, R.; Zhang, T.; Lin, S.; Chen, Z.; Zhao, B.; Li, H.; Wu, S. Relationship of Cardiovascular Disease Risk Factors and Noncoding RNAs with Hypertension: A Case-Control Study. BMC Cardiovasc. Disord. 2018, 18, 58.
- Wang, K.; Liu, F.; Liu, C.-Y.; An, T.; Zhang, J.; Zhou, L.-Y.; Wang, M.; Dong, Y.-H.; Li, N.; Gao, J.-N.; et al. The Long Noncoding RNA NRF Regulates Programmed Necrosis and Myocardial Injury during Ischemia and Reperfusion by Targeting miR-873. Cell Death Differ. 2016, 23, 1394–1405.
- Sun, F.; Zhuang, Y.; Zhu, H.; Wu, H.; Li, D.; Zhan, L.; Yang, W.; Yuan, Y.; Xie, Y.; Yang, S.; et al. LncRNA PCFL Promotes Cardiac Fibrosis via miR-378/GRB2 Pathway Following Myocardial Infarction. J. Mol. Cell. Cardiol. 2019, 133, 188–198.
- Ballantyne, M.D.; Pinel, K.; Dakin, R.S.; Vesey, A.T.; Diver, L.; MacKenzie, R.M.; Garcia, R.; Welsh, P.; Sattar, N.A.; Hamilton, G.; et al. Smooth Muscle Enriched Long Noncoding RNA (SMILR) Regulates Cell Proliferation. Circulation 2016, 133, 2050–2065.
- Huang, S.; Tao, W.; Guo, Z.; Cao, J.; Huang, X. Suppression of Long Noncoding RNA TTTY15 Attenuates Hypoxia-Induced Cardiomyocytes Injury by Targeting miR-455-5p. Gene 2019, 701, 1–8.
- Chen, J.; Hu, Q.; Zhang, B.-F.; Liu, X.-P.; Yang, S.; Jiang, H. Long Noncoding RNA UCA1 Inhibits Ischaemia/Reperfusion Injury Induced Cardiomyocytes Apoptosis via Suppression of Endoplasmic Reticulum Stress. Genes Genom. 2019, 41, 803–810.
- Yan, Y.; Zhang, B.; Liu, N.; Qi, C.; Xiao, Y.; Tian, X.; Li, T.; Liu, B. Circulating Long Noncoding RNA UCA1 as a Novel Biomarker of Acute Myocardial Infarction. BioMed Res. Int. 2016, 2016, 1–7.
- Zhang, J.; Yu, L.; Xu, Y.; Liu, Y.; Li, Z.; Xue, X.; Wan, S.; Wang, H. Long Noncoding RNA Upregulated in Hypothermia Treated Cardiomyocytes Protects against Myocardial Infarction through Improving Mitochondrial Function. Int. J. Cardiol. 2018, 266, 213–217.
- Micheletti, R.; Plaisance, I.; Abraham, B.J.; Sarre, A.; Ting, C.-C.; Alexanian, M.; Maric, D.; Maison, D.; Nemir, M.; Young, R.A.; et al. The Long Noncoding RNA Wisper Controls Cardiac Fibrosis and Remodeling. Sci. Transl. Med. 2017, 9, eaai9118.
- Jiao, L.; Li, M.; Shao, Y.; Zhang, Y.; Gong, M.; Yang, X.; Wang, Y.; Tan, Z.; Sun, L.; Xuan, L.; et al. lncRNA-ZFAS1 Induces Mitochondria-Mediated Apoptosis by Causing Cytosolic Ca2+ Overload in Myocardial Infarction Mice Model. Cell Death Dis. 2019, 10, 1–12.
- Yeh, C.-F.; Chang, Y.-C.E.; Lu, C.-Y.; Hsuan, C.-F.; Chang, W.-T.; Yang, K.-C. Expedition to the Missing Link: Long Noncoding RNAs in Cardiovascular Diseases. J. Biomed. Sci. 2020, 27, 1–16.
- Uchida, S.; Dimmeler, S. Long Noncoding RNAs in Cardiovascular Diseases. Circ. Res. 2015, 116, 737–750.
- Greco, S.; Somoza, A.S.; Devaux, Y.; Martelli, F. Long Noncoding RNAs and Cardiac Disease. Antioxidants Redox Signal. 2018, 29, 880–901.
- Hobuß, L.; Bär, C.; Thum, T. Long Non-Coding RNAs: At the Heart of Cardiac Dysfunction? Front. Physiol. 2019, 10, 30.
- Fang, Y.; Xu, Y.; Wang, R.; Hu, L.; Guo, D.; Xue, F.; Guo, W.; Zhang, D.; Hu, J.; Li, Y.; et al. Recent Advances on the Roles of LncRNAs in Cardiovascular Disease. J. Cell. Mol. Med. 2020, 24, 12246–12257.
- Collins, L.; Binder, P.; Chen, H.; Wang, X. Regulation of Long Non-Coding RNAs and MicroRNAs in Heart Disease: Insight into Mechanisms and Therapeutic Approaches. Front. Physiol. 2020, 11, 798.
- Viereck, J.; Thum, T. Circulating Noncoding RNAs as Biomarkers of Cardiovascular Disease and Injury. Circ. Res. 2017, 120, 381–399.
- Poller, W.; Dimmeler, S.; Heymans, S.; Zeller, T.; Haas, J.; Karakas, M.; Leistner, D.; Jakob, P.; Nakagawa, S.; Blankenberg, S.; et al. Non-Coding RNAs in Cardiovascular Diseases: Diagnostic and Therapeutic Perspectives. Eur. Hear. J. 2017, 39, 2704–2716.
- Moldovan, L.; Batte, K.E.; Trgovcich, J.; Wisler, J.; Marsh, C.B.; Piper, M. Methodological Challenges in Utilizing Mi RNAs as Circulating Biomarkers. J. Cell. Mol. Med. 2014, 18, 371–390.
- Kitow, J.; Derda, A.A.; Beermann, J.; Kumarswarmy, R.; Pfanne, A.; Fendrich, J.; Lorenzen, J.M.; Xiao, K.; Bavendiek, U.; Bauersachs, J.; et al. Mitochondrial Long Noncoding RNAs as Blood Based Biomarkers for Cardiac Remodeling in Patients with Hypertrophic Cardiomyopathy. Am. J. Physiol. Heart Circ. Physiol. 2016, 311, 707–712.
- Li, D.; Chen, G.; Yang, J.; Fan, X.; Gong, Y.; Xu, G.; Cui, Q.; Geng, B. Transcriptome Analysis Reveals Distinct Patterns of Long Noncoding RNAs in Heart and Plasma of Mice with Heart Failure. PLoS ONE 2013, 8, e77938.
- Goretti, E.; Wagner, D.R.; Devaux, Y. Mirnas as Biomarkers of Myocardial Infarction: A Step Forward towards Personalized Medicine? Trends Mol. Med. 2014, 20, 716–725.
- Congrains, A.; Kamide, K.; Oguro, R.; Yasuda, O.; Miyata, K.; Yamamoto, E.; Kawai, T.; Kusunoki, H.; Yamamoto, H.; Takeya, Y.; et al. Genetic Variants at the 9p21 Locus Contribute to Atherosclerosis through Modulation of ANRIL and CDKN2A/B. Atherosclerosis 2012, 220, 449–455.
- Iacobucci, I.; Sazzini, M.; Garagnani, P.; Ferrari, A.; Boattini, A.; Lonetti, A.; Papayannidis, C.; Mantovani, V.; Marasco, E.; Ottaviani, E.; et al. A Polymorphism in the Chromosome 9p21 ANRIL Locus Is Associated to Philadelphia Positive Acute Lymphoblastic Leukemia. Leuk. Res. 2011, 35, 1052–1059.
- Diederichs, S. The Four Dimensions of Noncoding RNA Conservation. Trends Genet. 2014, 30, 121–123.
- Spitale, R.C.; Crisalli, P.; Flynn, R.A.; Torre, E.A.; Kool, E.T.; Chang, H.Y. RNA SHAPE Analysis in Living Cells. Nat. Chem. Biol. 2013, 9, 18–20.
- Ding, Y.; Tang, Y.; Kwok, C.K.; Zhang, Y.; Bevilacqua, P.C.; Assmann, S.M. In Vivo Genome-Wide Profiling of RNA Secondary Structure Reveals Novel Regulatory Features. Nature 2014, 505, 696–700.
- Das, S.; Shah, R.; Dimmeler, S.; Freedman, J.E.; Holley, C.; Lee, J.-M.; Moore, K.; Musunuru, K.; Wang, D.-Z.; Xiao, J.; et al. Noncoding RNAs in Cardiovascular Disease: Current Knowledge, Tools and Technologies for Investigation, and Future Directions: A Scientific Statement from the American Heart Association. Circ. Genom. Precis. Med. 2020, 13.
- Volders, P.-J.; Anckaert, J.; Verheggen, K.; Nuytens, J.; Martens, L.; Mestdagh, P.; Vandesompele, J. LNCipedia 5: Towards a Reference Set of Human Long Non-Coding RNAs. Nucleic Acids Res. 2019, 47, 135–139.
- Li, J.; Ma, W.; Zeng, P.; Wang, J.; Geng, B.; Yang, J.; Cui, Q. LncTar: A Tool for Predicting the RNA Targets of Long Noncoding RNAs. Brief. Bioinform. 2014, 16, 806–812.
- Ma, L.; Li, A.; Zou, D.; Xu, X.; Xia, L.; Yu, J.; Bajic, V.B.; Zhang, Z. LncRNAWiki: Harnessing Community Knowledge in Collaborative Curation of Human Long Non-Coding RNAs. Nucleic Acids Res. 2014, 43, 187–192.
- Ma, L.; Cao, J.; Liu, L.; Li, Z.; Shireen, H.; Pervaiz, N.; Batool, F.; Raza, R.Z.; Zou, D.; Bao, Y.; et al. Community Curation and Expert Curation of Human Long Noncoding RNAs with LncRNAWiki and LncBook. Curr. Protoc. Bioinform. 2019, 67, e82.
