Intrinsically disordered proteins (IDPs) are highly prevalent and play important roles in biology and human diseases. It is now also recognized that many IDPs remain dynamic even in specific complexes and functional assemblies. Computer simulations are essential for deriving a molecular description of the disordered protein ensembles and dynamic interactions for a mechanistic understanding of IDPs in biology, diseases, and therapeutics.
- conformational ensemble
- enhanced sampling
- generalized Born
- Gō-model
- implicit solvent
- liquid-liquid phase transition
- replica exchange
- protein force fields
1. Introduction
Intrinsically disordered proteins (IDPs) or regions (IDRs), compared to well-structured proteins, do not have stable tertiary structures under physiological conditions. Nevertheless, IDPs or IDRs can be found in nearly a third of proteins encoded in the human proteome [1], and they play key roles in a variety of biological processes that underlie vital cellular functions ranging from signaling and regulation to transport [2][3]. The inherent thermodynamic instability of an IDP’s conformation allows it to respond sensitively to numerous stimuli, including binding, changes in cellular environments (e.g., pH), and post-translational modifications [4][5][6][7][8]. Such conformational plasticity arguably enables IDPs to interact with multiple signaling pathways and serve as scaffolds to form multi-protein complexes [9]. Importantly, IDPs and IDRs house around 25% of disease-associated missense mutations [10]. They have been considered promising therapeutic targets for treating various diseases (such as chronic diseases) [11][12][13]. While many IDPs have been shown to undergo binding-induced folding transitions upon specific binding [3], many examples are also emerging to demonstrate that IDPs can remain unstructured even in specific complexes and functional assemblies [14][15][16][17][18][19][20]. Such a dynamic mode of specific protein interactions seems much more prevalent than previously thought [21][22][23].
It is very challenging to provide reliable descriptions of the conformational ensembles of IDPs and IDRs. A disordered state does not lend itself to traditional structural determination methods that are geared toward describing a coherent set of similar structures. Biophysical techniques, such as NMR, SAXS, and FRET, can provide complementary information on various local and long-range structural organizations [7]. However, these ensemble-averaged measurements alone are not sufficient to unambiguously define the heterogeneous ensemble, due to the severely underdetermined nature of the structure calculation problem [8][24][25]. As a result, studies of IDPs have relied heavily in the traditional structure-function paradigm, by solving the folded structure of the bound state, analyzing coupled binding and folding mechanisms, or identifying putative pre-existing functional structures in the unbound state [3]. However, the disordered ensemble itself is arguably the central conduit of cellular signaling. The functional mechanism of an IDP is encoded in how the disordered ensemble as a whole responds to various stimuli, be it cooperative binding-induced folding or the redistribution of conformational sub-states in dynamic interactions. Multiple cellular signals can be naturally integrated through cooperative responses of the whole dynamic ensemble [26][27][28]. Therefore, there is a critical need for reliable characterization of disordered protein conformation ensembles, in both bound and unbound states, in order to establish the molecular basis of IDPs and IDRs in various physiological and pathophysiological processes.
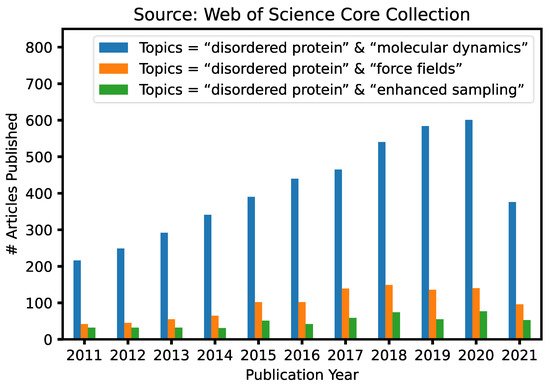
Given the fundamental challenges of characterizing disordered protein states based on ensemble-averaged measurements alone, molecular modeling and simulations have a crucial and unique role to play in mechanistic studies of IDPs and IDRs [29][30][31][32][33]. This is reflected in continuously increasing numbers of research articles that contain keywords “intrinsically disordered” and “molecular dynamics” published in the last 10 years ( Figure 1 ). A particularly attractive approach is to first generate the disordered ensemble using transferable, physics-based force fields without any experimental restraints and then use the later for independent validation [7]. Such de novo simulations of disordered protein ensembles require both high force field accuracy and adequate sampling of relevant conformational space, pushing the limit of these two central ingredients of molecular dynamics (MD) and Monte Carlo (MC) simulations. The challenges of simulating disordered proteins have driven significant interest in developing better protein force fields and advanced sampling methods ( Figure 1 ). In particular, important advances have been made in the state-of-the-art atomistic force fields for describing the conformational equilibria of ordered and disordered proteins [13]. Enhanced sampling techniques have played crucial roles in both the development and application of atomistic force fields, by allowing one to cross energy barriers faster and accelerate the conformational sampling of IDPs [34][35][36][37][38][39][40][41]. Nonetheless, atomistic simulations still have limited capability in describing large systems such as biological condensates [42]. For this, multi-scale approaches are necessary to bridge the gaps in experimental and computational time- and length-scales, including implicit solvent models, which remove the solvent degrees of freedom [8], and various coarse-grained models, which significantly reduce both proteins and solvent degrees of freedom [43].
2. Challenges of Simulating IDP Conformational Equilibria
Compared to the globular proteins that have one or a few well-defined global energy minima, the energy landscape of an IDP is flatter and generally includes many local energy minima separated by modest energy barriers [44]. IDPs and IDRs typically have fewer hydrophobic residues, but a larger number of polar or charged as well as disorder-promoting residues (such as glycine and proline) [45]. These sequence features hamper the formation of hydrophobic cores that drive protein folding and thus prevent the formation of stable tertiary structures. Instead, IDPs and IDRs favor forming an ensemble of unfolded or partially folded states. This presents a major challenge for simulation and depends critically on the ability of the force fields to accurately describe the energetics of relevant conformational states, especially for capturing both folded and unfolded states of an IDP. For example, one recent study tested atomistic simulations of IDPs for eight force fields and found marked differences in the describing the conformational ensembles of IDPs, in particular the secondary structure content [46]. Similar observations have also been made in other benchmark studies, consistently showing that protein force fields previously optimized for folded proteins are not suitable for simulating disordered protein states, largely due to over-stabilization of protein-protein interactions [47]. These benchmark studies also suggested that the key towards better protein force field was to rebalance protein–protein, protein–water, and water–water interactions.
Besides accurate force fields, reliable simulation of IDPs also hinges on sufficient sampling of many relevant conformation states within a reasonable simulation time. Standard MD simulations are generally insufficient to generate representative conformational ensembles, even using the most accurate protein force fields coupled with advance of GPU computing or specialized hardware such as the ANTON supercomputer [48]. For example, a recent reanalysis of a 30-μs ANTON trajectory of a 40-residue Aβ40 peptide in explicit solvent revealed very limited convergence even at the secondary structure level [13]. This can be attributed to the diverse and large accessible conformational space of an IDP and the potentially high free energy barriers separating various sub-states that require exponentially longer time to cross. Note that typical simulation times on conventional hardware (such as GPUs) are at least one-order of magnitude shorter. There is thus great danger in relying on standard MD to calculate disordered protein conformational ensembles at the atomistic level. There is a critical need to develop and leverage so-called enhanced sampling techniques, which aim to generate statistically meaningful conformational ensembles with dramatically less computation.
Computational studies of IDP interaction and assembly are even more demanding. The conformational equilibrium of an IDP can respond sensitively to specific and nonspecific binding, potentially shifting from a disordered to somewhat ordered state or fully folded state. In principle, simulations could provide the much-needed spatial and time resolutions to elucidate the kinetics and thermodynamics of coupled folding and binding processes and characterize the mechanistic features. However, the challenge is that this coupled process of folding and binding is a complex reaction involving the formation of many noncovalent interactions, which requires extremely long simulations generally beyond the current capabilities at the atomistic level. As such, coarse-grained models are generally required for computational studies of IDP interaction and assembly.
3. The State-of-the-Art Protein Force Fields for Describing IDP Conformations
Empirical protein force fields are potential energy functions that typically include physics-motivated bonded and non-bonded terms carefully parameterized based on a wide range of theoretical and experimental data [49]. These force fields can in principle be transferable between folded proteins and IDPs. To achieve this, it is also critical to develop suitable water models and better describe the water–protein interactions [50][51]. Two recent review articles have already provided comprehensive descriptions on the latest development of better protein force fields [49][52]. We therefore briefly summarize the state-of-the-art of nonpolarizable and polarizable force fields for IDP dynamics and interactions.
Many previous nonpolarizable force fields have significant shortcomings for describing the unfolded or disordered proteins. For example, they typically provide a poor description of the secondary structure content for IDPs and have a preference to give too compact conformations with respect to the experimentally measured dimension of IDPs [46][53]. These problems were likely attributed to the unbalanced parameterization of dihedral torsion space and the description of protein–protein and protein–water interactions [54]. As a result, most of the improved force fields managed to give more accurate secondary structure propensities by adjusting dihedral parameters or adding grid-based energy correction map (CMAP) parameters [52]. The over-compactness of disordered proteins can be alleviated by modifying protein–water van der Waals interactions or combining with refined water models [50]. Representative state-of-the-art force fields includes the latest CHARMM36m/TIP3P* [55], ff19SB/OPC [56], and a99SB-disp/TIP4P-D [48]. Many benchmark studies have consistently demonstrated that these refined force fields do provide significant improvements in describing not only single folded and disordered proteins, but also the multiprotein systems that are either soluble or aggregate in the solution [53][57][58][59][60]. At the same time, these studies also identified significant remaining limitations in the description of the noncovalent interactions in the multiprotein systems [58]. Recognizing limitations in the ability of a99SB-disp/TIP4P-D force field to accurately describe the protein–protein interactions, a new force field, DES-Amber, was recently developed to provide more accurate simulations of protein–protein complexes while maintaining reliable descriptions of both ordered and disordered single-chain proteins [59]. However, DES-Amber is still limited in reproducing the experimental protein–protein association free energies of some protein complexes, in particular for the systems with highly polar interfaces [59]. In the latter case, it was found that the charged sidechains were buried at the protein–protein interface instead of being solvent-exposed. It was further suggested that nonpolarizable force fields were fundamentally limited in achieving a balanced description of charged groups that were solvent-exposed or buried at a protein–protein interface.
Polarizable force fields explicitly consider the electronic polarization using various empirical models to provide better description of charged and polar protein motifs in heterogeneous biomolecular environments [61]. Exciting progress has been made in the last few years and several polarizable force fields are now available for the stable simulation of proteins in both aqueous and membrane environments [62][63]. Simulations using the latest polarizable force fields have also showed a high level of consistency with experimental observations, particularly the ion solvation and binding thermodynamics, permeation free energy of ions or small charged molecules into the cell membrane, and protein–ligand binding [61]. For example, the Drude-2013 polarizable force field, compared to CHARMM36 force field, is more accurate in describing the folding cooperativity of (AAQAA) 3 peptide, which can be attributed to enhanced backbone dipole moments in the helix state [64]. Additional studies are still needed to show the necessity of considering polarizable force fields in IDP simulations, where the significantly higher computational cost adds to the challenge of generating converged ensembles [61]. Existing comparisons suggest that polarizable force fields, including AMOEBA and Drude models, still frequently have problems in reproducing the nature structures and folding of proteins [65][66][67]. For example, stronger protein–water interactions in polarizable force fields can destabilize the native protein structure, in opposition to the observations from nonpolarizable force fields where protein–water interactions have traditionally been underestimated [42]. Nonetheless, it can be anticipated that polarizable force fields will continue to be improved and become increasingly important for simulating IDP structure and interactions.
4. Enhanced Sampling Methods for Sampling IDP Conformational Ensembles
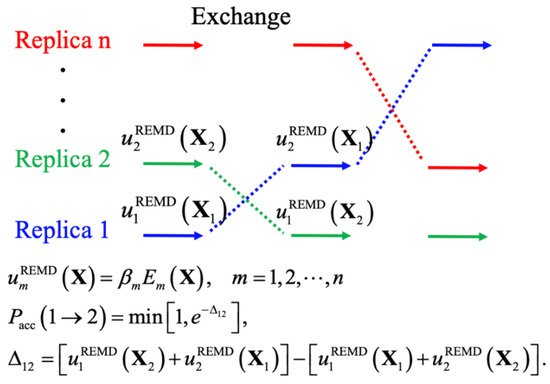
Enhanced sampling techniques generally accelerate the crossing of energy barriers to achieve better sampling efficiency, such as by introducing bias potentials, modifying the potential energy itself, and changing the effective temperature. These techniques have proven essential in atomistic simulations of IDPs [68][69], yielding levels of convergence that could not be achieved even with drastically longer standard constant-temperature MD simulations [13]. The central idea of biased MD simulations is similar to importance sampling in MC simulations, where a biased potential is introduced to construct a flat free energy landscape along single or multiple collective variables of interest, such that many states can be readily sampled due to the removal of free energy barriers. The replica-exchange (REX) class of sampling methods, particularly replica exchange molecular dynamics (REMD), has been one of the most popular methods for simulating protein conformations. Figure 2 shows the general scheme of REMD simulations, where the key point is to first set up multiple replicas with different unitless unbiased or biased potentials, given as the energy over k B T ( T is the temperature), and then use the Metropolis rule to allow MC to exchange the replicas and maintain the detailed balance. A key advantage of using multiple replicas and maintaining detailed balance is avoiding the reweighting problem generally required for biased simulations. Note that virtually all biased sampling strategies can be readily incorporated within the REX framework to benefit from both classes of enhanced sampling, including metadynamics (MTD) [70][71], accelerated MD (aMD) [72], umbrella sampling (US) [73][74], and integrated tempering sampling [75]. In practice, effective REMD protocols require a proper choice of (1) the optimal number of replicas and proper distributions of conditions, to ensure a uniform exchange acceptance rate and efficient random walk in the condition space, and (2) the choice of those unitless (biased) potentials for effective conformational diffusion at each condition [76]. Here, we divide various enhanced sampling strategies into two general groups depending on the need for collective variables and discuss their recent applications to IDP conformational sampling. These methods are summarized Table 1 .
| Types | Sampling Methods | Key Features | References |
|---|---|---|---|
| CV-based | WT-MTD | History-based adaptive bias potentials | [70][71] |
| Bias-exchange MTD | Multiple replicas with bias on different CVs | [77] | |
| Umbrella sampling | Pre-determined bias potentials | [78] | |
| Machine learning | On-the-fly discover optimal CVs | [79][80] | |
| Tempering-based | Simulated tempering | Random walk in the temperature space | [81] |
| Parallel tempering | Multiple replicas to avoid the need for estimating the density of states | [36] | |
| Integrated tempering | Integral of Boltzmann distributions over a range of temperatures as the bias | [75] | |
| Solute tempering | Scaling the energies of only selected atoms or terms to achieve effective tempering | [37][82][83] | |
| Accelerated MD | GaMD | Boost potentials to accelerate barrier crossing | [84] |
| Combinations | MSES | Temperature/Hamiltonian replica exchange simulation by coupling CG and atomistic models | [34] |
| REUS/REST | Combined REUS and REST | [85] | |
| REUS/GaMD | Combined REUS and GaMD | [86] | |
| Integrated aMD | Integrated aMD and integrated tempering | [67][87] | |
| PT-MTD | Combined the WT-MTD with PT | [77] |
MTD and its variants have been considered one of the most important collective variables (CV)-based sampling methods for protein simulations [88]. MTD uses a history-dependent bias potential, which is generally a sum of Gaussians, to eventually construct a flat free energy landscape along the predetermined CV(s). A well-tempered MTD (WT-MTD) was later developed to increase the convergence, by gradually reducing the size of Gaussians based on the total accumulated bias potential [70][71]. Furthermore, the parallel tempering MTD (PT-MTD) and the combinations with other biased sampling methods have been also developed to increase the sampling efficiency and convergence of free energy calculations [89][90]. Representative examples include the PT-MTD that combines WT-MTD with PT or bias-exchange MTD that uses a different CV in each replica, rather than exchanging the temperatures. For example, the PT-WTD and bias-exchange MTD has been employed to obtain the conformational ensembles and coupled binding and folding of disordered pKID and KID proteins, using the α-score of helical structures as CVs [77]. It has also been shown that the REMD-based MTD, compared to conventional MTD or T-REMD, can enhance the conformational sampling of N-Glycans using dihedral angles as CVs to characterize the global motions [91]. The binding mechanism of two disordered peptides, NRF2 and PTMA, was simulated by the WT-MTD, and the results showed that the WT-MTD method could provide converged free energy profiles with 1.5 μs of sampling time [92]. Together, these applications have shown that MTD-class of sampling methods can be effectively applied to IDP simulations. Beside MTD, another important class of CV-based sampling strategy is the US method [74]. US is not strictly an enhanced sampling method like MTD. It typically uses multiple harmonic potentials to focus on sampling various states along the collective variables of interest. US is often combined with REMD in studies of IDPs, as illustrated in a recent 2D window-exchange US simulation of the coupled folding and binding mechanism of HdeA homodimer [78]. The simulation was able to capture rare unfolding transitions of the dimer at neutral pH and provided a detailed description of the transition pathways.
REST has proven to be one of the most reliable choices for enhanced sampling of protein folding and particularly disordered conformational ensembles [93][94]. Sugita and co-workers leveraged gREST to target the dihedral-angle energy term and successfully sampled folding transitions of beta-hairpins and Trp-cage in explicit water, using fewer replicas but covering wider conformational space compared to REST2 [82]. Walsh et al. applied REST to investigate n16N disordered peptide conformational ensembles [95]. The conformations obtained via REST methods showed a high consistency with NMR experimental data. Furthermore, REST are specifically appropriate in simulating IDRs as the disordered region can be targeted in REST without tempering the well-structured region (or water). Zhou and co-workers studied the disordered loop of Staphylococcus aureus sortase A (SrtA) to order transition upon binding to calcium [96]. Chen and Liu characterized Bcl-xL interfacial conformational dynamics in explicit solvent [97]. Both works directly showed that REST covered broader conformational spaces for intrinsically disordered regions and led to faster convergence compared to either standard MD or T-REMD simulations. REST simulations have also been successfully integrated with experiments to study how cancer-associated mutations and drug molecules may modulate the disordered ensembles of p53-TAD and Aβ peptides in recent years [98][99][100][101].
Despite the success of REST for CV-free enhanced sampling, it does not benefit from targeted acceleration along specific CVs that are known to be rate limiting. For this, REST (or REX in general) has been combined with CV-based enhanced sampling to maximize the efficiency of sampling the complex, high dimensional conformational space of proteins. Some of the examples are discussed in the sections above. Here, we note a couple additional recent examples. By integrating free energy perturbation (FEP) and REST methods, Abel et al. obtained more thorough samplings of different ligand conformations around the active site and realized relative binding affinity predictions [102]. Okamoto and co-workers have applied the REUS/REST two-dimensional replica-exchange method to predict two protein–ligand complex systems with the help of REST to weaken the solute–solvent interactions but improve the binding events and REUS to enhance the sampling along with the reaction coordinates [85].
References
- Csizmok, V.; Follis, A.V.; Kriwacki, R.W.; Forman-Kay, J.D. Dynamic Protein Interaction Networks and New Structural Paradigms in Signaling. Chem. Rev. 2016, 116, 6424–6462.
- Oldfield, C.J.; Dunker, A.K. Intrinsically Disordered Proteins and Intrinsically Disordered Protein Regions. Annu. Rev. Biochem. 2014, 83, 553–584.
- Wright, P.E.; Dyson, H.J. Intrinsically disordered proteins in cellular signalling and regulation. Nat. Rev. Mol. Cell Biol. 2015, 16, 18–29.
- Uversky, V.N. Intrinsically disordered proteins and their (disordered) proteomes in neurodegenerative disorders. Front. Aging Neurosci. 2015, 7, 18.
- Dyson, H.J.; Wright, P.E. Intrinsically unstructured proteins and their functions. Nat. Rev. Mol. Cell Biol. 2005, 6, 197–208.
- Owen, I.; Shewmaker, F. The Role of Post-Translational Modifications in the Phase Transitions of Intrinsically Disordered Proteins. Int. J. Mol. Sci. 2019, 20, 5501.
- Chen, J. Towards the physical basis of how intrinsic disorder mediates protein function. Arch. Biochem. Biophys. 2012, 524, 123–131.
- Das, R.K.; Ruff, K.M.; Pappu, R.V. Relating sequence encoded information to form and function of intrinsically disordered proteins. Curr. Opin. Struct. Biol. 2015, 32, 102–112.
- Hatos, A.; Hajdu-Soltesz, B.; Monzon, A.M.; Palopoli, N.; Alvarez, L.; Aykac-Fas, B.; Bassot, C.; Benitez, G.I.; Bevilacqua, M.; Chasapi, A.; et al. DisProt: Intrinsic protein disorder annotation in 2020. Nucleic Acids Res. 2020, 48, D269–D276.
- Vacic, V.; Iakoucheva, L.M. Disease mutations in disordered regions—Exception to the rule? Mol. Biosyst. 2012, 8, 27–32.
- Kulkarni, P.; Uversky, V.N. Intrinsically Disordered Proteins in Chronic Diseases. Biomolecules 2019, 9, 147.
- Oldfield, C.J.; Cheng, Y.; Cortese, M.S.; Brown, C.J.; Uversky, V.N.; Dunker, A.K. Comparing and Combining Predictors of Mostly Disordered Proteins. Biochemistry 2005, 44, 1989–2000.
- Chen, J.; Liu, X.; Chen, J. Targeting Intrinsically Disordered Proteins through Dynamic Interactions. Biomolecules 2020, 10, 743.
- Mittag, T.; Marsh, J.; Grishaev, A.; Orlicky, S.; Lin, H.; Sicheri, F.; Tyers, M.; Forman-Kay, J.D. Structure/Function Implications in a Dynamic Complex of the Intrinsically Disordered Sic1 with the Cdc4 Subunit of an SCF Ubiquitin Ligase. Structure 2010, 18, 494–506.
- McDowell, C.; Chen, J.; Chen, J. Potential Conformational Heterogeneity of p53 Bound to S100B(betabeta). J. Mol. Biol. 2013, 425, 999–1010.
- Wu, H.; Fuxreiter, M. The Structure and Dynamics of Higher-Order Assemblies: Amyloids, Signalosomes, and Granules. Cell 2016, 165, 1055–1066.
- Krois, A.S.; Ferreon, J.C.; Martinez-Yamout, M.A.; Dyson, H.J.; Wright, P.E. Recognition of the disordered p53 transactivation domain by the transcriptional adapter zinc finger domains of CREB-binding protein. Proc. Natl. Acad. Sci. USA 2016, 113, E1853–E1862.
- Csizmok, V.; Orlicky, S.; Cheng, J.; Song, J.; Bah, A.; Delgoshaie, N.; Lin, H.; Mittag, T.; Sicheri, F.; Chan, H.S.; et al. An allosteric conduit facilitates dynamic multisite substrate recognition by the SCFCdc4 ubiquitin ligase. Nat. Commun. 2017, 8, 13943.
- Borgia, A.; Borgia, M.B.; Bugge, K.; Kissling, V.M.; Heidarsson, P.O.; Fernandes, C.B.; Sottini, A.; Soranno, A.; Buholzer, K.J.; Nettels, D.; et al. Extreme disorder in an ultrahigh-affinity protein complex. Nature 2018, 555, 61–66.
- Clark, S.; Myers, J.B.; King, A.; Fiala, R.; Novacek, J.; Pearce, G.; Heierhorst, J.; Reichow, S.L.; Barbar, E.J. Multivalency regulates activity in an intrinsically disordered transcription factor. Elife 2018, 7, e36258.
- Fuxreiter, M. Fuzziness in Protein Interactions-A Historical Perspective. J. Mol. Biol. 2018, 430, 2278–2287.
- Weng, J.; Wang, W. Dynamic multivalent interactions of intrinsically disordered proteins. Curr. Opin. Struct. Biol. 2019, 62, 9–13.
- Miskei, M.; Antal, C.; Fuxreiter, M. FuzDB: Database of fuzzy complexes, a tool to develop stochastic structure-function relationships for protein complexes and higher-order assemblies. Nucleic Acids Res. 2017, 45, D228–D235.
- Ganguly, D.; Chen, J. Structural interpretation of paramagnetic relaxation enhancement-derived distances for disordered protein states. J. Mol. Biol. 2009, 390, 467–477.
- Fisher, C.K.; Stultz, C.M. Constructing ensembles for intrinsically disordered proteins. Curr. Opin. Struct. Biol. 2011, 21, 426–431.
- Ferreon, A.C.; Ferreon, J.C.; Wright, P.E.; Deniz, A.A. Modulation of allostery by protein intrinsic disorder. Nature 2013, 498, 390–394.
- Garcia-Pino, A.; Balasubramanian, S.; Wyns, L.; Gazit, E.; De Greve, H.; Magnuson, R.D.; Charlier, D.; van Nuland, N.A.; Loris, R. Allostery and intrinsic disorder mediate transcription regulation by conditional cooperativity. Cell 2010, 142, 101–111.
- Berlow, R.B.; Dyson, H.J.; Wright, P.E. Expanding the Paradigm: Intrinsically Disordered Proteins and Allosteric Regulation. J. Mol. Biol. 2018, 430, 2309–2320.
- Levine, Z.A.; Shea, J.-E. Simulations of disordered proteins and systems with conformational heterogeneity. Curr. Opin. Struct. Biol. 2017, 43, 95–103.
- Knott, M.; Best, R.B. A preformed binding interface in the unbound ensemble of an intrinsically disordered protein: Evidence from molecular simulations. PLoS Comput. Biol. 2012, 8, e1002605.
- Mao, A.H.; Crick, S.L.; Vitalis, A.; Chicoine, C.L.; Pappu, R.V. Net charge per residue modulates conformational ensembles of intrinsically disordered proteins. Proc. Natl. Acad. Sci. USA 2010, 107, 8183–8188.
- Ganguly, D.; Chen, J. Atomistic details of the disordered states of KID and pKID. implications in coupled binding and folding. J. Am. Chem. Soc. 2009, 131, 5214–5223.
- Zhang, W.; Ganguly, D.; Chen, J. Residual structures, conformational fluctuations, and electrostatic interactions in the synergistic folding of two intrinsically disordered proteins. PLoS Comput. Biol. 2012, 8, e1002353.
- Zhang, W.; Chen, J. Accelerate Sampling in Atomistic Energy Landscapes Using Topology-Based Coarse-Grained Models. J. Chem. Theory Comput. 2014, 10, 918–923.
- Moritsugu, K.; Terada, T.; Kidera, A. Scalable free energy calculation of proteins via multiscale essential sampling. J. Chem. Phys. 2010, 133, 224105.
- Sugita, Y.; Okamoto, Y. Replica-exchange molecular dynamics method for protein folding. Chem. Phys. Lett. 1999, 314, 141–151.
- Liu, P.; Kim, B.; Friesner, R.A.; Berne, B.J. Replica exchange with solute tempering: A method for sampling biological systems in explicit water. Proc. Natl. Acad. Sci. USA 2005, 102, 13749–13754.
- Mittal, A.; Lyle, N.; Harmon, T.S.; Pappu, R.V. Hamiltonian Switch Metropolis Monte Carlo Simulations for Improved Conformational Sampling of Intrinsically Disordered Regions Tethered to Ordered Domains of Proteins. J. Chem. Theory Comput. 2014, 10, 3550–3562.
- Peter, E.K.; Shea, J.E. A hybrid MD-kMC algorithm for folding proteins in explicit solvent. Phys. Chem. Chem. Phys. 2014, 16, 6430–6440.
- Zhang, C.; Ma, J. Enhanced sampling and applications in protein folding in explicit solvent. J. Chem. Phys. 2010, 132, 244101.
- Zheng, L.Q.; Yang, W. Practically Efficient and Robust Free Energy Calculations: Double-Integration Orthogonal Space Tempering. J. Chem. Theory Comput. 2012, 8, 810–823.
- Best, R.B. Computational and theoretical advances in studies of intrinsically disordered proteins. Curr. Opin. Struct. Biol. 2017, 42, 147–154.
- Kmiecik, S.; Gront, D.; Kolinski, M.; Wieteska, L.; Dawid, A.E.; Kolinski, A. Coarse-Grained Protein Models and Their Applications. Chem. Rev. 2016, 116, 7898–7936.
- Arai, M. Unified understanding of folding and binding mechanisms of globular and intrinsically disordered proteins. Biophys. Rev. 2018, 10, 163–181.
- Bhattacharya, S.; Lin, X. Recent Advances in Computational Protocols Addressing Intrinsically Disordered Proteins. Biomolecules 2019, 9, 146.
- Rauscher, S.; Gapsys, V.; Gajda, M.J.; Zweckstetter, M.; de Groot, B.L.; Grubmüller, H. Structural Ensembles of Intrinsically Disordered Proteins Depend Strongly on Force Field: A Comparison to Experiment. J. Chem. Theory Comput. 2015, 11, 5513–5524.
- Best, R.B.; Zhu, X.; Shim, J.; Lopes, P.E.; Mittal, J.; Feig, M.; Mackerell, A.D., Jr. Optimization of the additive CHARMM all-atom protein force field targeting improved sampling of the backbone phi, psi and side-chain chi(1) and chi(2) dihedral angles. J. Chem. Theory Comput. 2012, 8, 3257–3273.
- Robustelli, P.; Piana, S.; Shaw, D.E. Developing a molecular dynamics force field for both folded and disordered protein states. Proc. Natl. Acad. Sci. USA 2018, 115, E4758–E4766.
- Huang, J.; MacKerell, A.D. Force field development and simulations of intrinsically disordered proteins. Curr. Opin. Struct. Biol. 2018, 48, 40–48.
- Piana, S.; Donchev, A.G.; Robustelli, P.; Shaw, D.E. Water Dispersion Interactions Strongly Influence Simulated Structural Properties of Disordered Protein States. J. Phys. Chem. B 2015, 119, 5113–5123.
- Wu, H.-N.; Jiang, F.; Wu, Y.-D. Significantly Improved Protein Folding Thermodynamics Using a Dispersion-Corrected Water Model and a New Residue-Specific Force Field. J. Phys. Chem. Lett. 2017, 8, 3199–3205.
- Mu, J.; Liu, H.; Zhang, J.; Luo, R.; Chen, H.-F. Recent Force Field Strategies for Intrinsically Disordered Proteins. J. Chem. Inf. Modeling 2021, 61, 1037–1047.
- Song, D.; Liu, H.; Luo, R.; Chen, H.-F. Environment-Specific Force Field for Intrinsically Disordered and Ordered Proteins. J. Chem. Inf. Modeling 2020, 60, 2257–2267.
- Yang, S.; Liu, H.; Zhang, Y.; Lu, H.; Chen, H. Residue-Specific Force Field Improving the Sample of Intrinsically Disordered Proteins and Folded Proteins. J. Chem. Inf. Modeling 2019, 59, 4793–4805.
- Huang, J.; Rauscher, S.; Nawrocki, G.; Ran, T.; Feig, M.; de Groot, B.L.; Grubmüller, H.; MacKerell, A.D. CHARMM36m: An improved force field for folded and intrinsically disordered proteins. Nat. Methods 2017, 14, 71–73.
- Tian, C.; Kasavajhala, K.; Belfon, K.A.A.; Raguette, L.; Huang, H.; Migues, A.N.; Bickel, J.; Wang, Y.; Pincay, J.; Wu, Q.; et al. ff19SB: Amino-Acid-Specific Protein Backbone Parameters Trained against Quantum Mechanics Energy Surfaces in Solution. J. Chem. Theory Comput. 2020, 16, 528–552.
- Rahman, M.U.; Rehman, A.U.; Liu, H.; Chen, H.-F. Comparison and Evaluation of Force Fields for Intrinsically Disordered Proteins. J. Chem. Inf. Modeling 2020, 60, 4912–4923.
- Abriata, L.A.; Dal Peraro, M. Assessment of transferable forcefields for protein simulations attests improved description of disordered states and secondary structure propensities, and hints at multi-protein systems as the next challenge for optimization. Comput. Struct. Biotechnol. J. 2021, 19, 2626–2636.
- Piana, S.; Robustelli, P.; Tan, D.; Chen, S.; Shaw, D.E. Development of a Force Field for the Simulation of Single-Chain Proteins and Protein–Protein Complexes. J. Chem. Theory Comput. 2020, 16, 2494–2507.
- Song, D.; Luo, R.; Chen, H.-F. The IDP-Specific Force Field ff14IDPSFF Improves the Conformer Sampling of Intrinsically Disordered Proteins. J. Chem. Inf. Modeling 2017, 57, 1166–1178.
- Jing, Z.; Liu, C.; Cheng, S.Y.; Qi, R.; Walker, B.D.; Piquemal, J.-P.; Ren, P. Polarizable Force Fields for Biomolecular Simulations: Recent Advances and Applications. Annu. Rev. Biophys. 2019, 48, 371–394.
- Bedrov, D.; Piquemal, J.-P.; Borodin, O.; MacKerell, A.D.; Roux, B.; Schröder, C. Molecular Dynamics Simulations of Ionic Liquids and Electrolytes Using Polarizable Force Fields. Chem. Rev. 2019, 119, 7940–7995.
- Inakollu, V.S.S.; Geerke, D.P.; Rowley, C.N.; Yu, H. Polarisable force fields: What do they add in biomolecular simulations? Curr. Opin. Struct. Biol. 2020, 61, 182–190.
- Huang, J.; MacKerell, A.D., Jr. Induction of Peptide Bond Dipoles Drives Cooperative Helix Formation in the (AAQAA)3 Peptide. Biophys. J. 2014, 107, 991–997.
- Kamenik, A.S.; Handle, P.H.; Hofer, F.; Kahler, U.; Kraml, J.; Liedl, K.R. Polarizable and non-polarizable force fields: Protein folding, unfolding, and misfolding. J. Chem. Phys. 2020, 153, 185102.
- Wang, A.; Zhang, Z.; Li, G. Higher Accuracy Achieved in the Simulations of Protein Structure Refinement, Protein Folding, and Intrinsically Disordered Proteins Using Polarizable Force Fields. J. Phys. Chem. Lett. 2018, 9, 7110–7116.
- Wang, A.; Peng, X.; Li, Y.; Zhang, D.; Zhang, Z.; Li, G. Quality of force fields and sampling methods in simulating pepX peptides: A case study for intrinsically disordered proteins. Phys. Chem. Chem. Phys. 2021, 23, 2430–2437.
- Yang, Y.I.; Shao, Q.; Zhang, J.; Yang, L.; Gao, Y.Q. Enhanced sampling in molecular dynamics. J. Chem. Phys. 2019, 151, 070902.
- Wang, A.H.; Zhang, Z.C.; Li, G.H. Advances in Enhanced Sampling Molecular Dynamics Simulations for Biomolecules. Chin. J. Chem. Phys. 2019, 32, 277–286.
- Barducci, A.; Bonomi, M.; Parrinello, M. Metadynamics. WIREs Comput. Mol. Sci. 2011, 1, 826–843.
- Barducci, A.; Bussi, G.; Parrinello, M. Well-Tempered Metadynamics: A Smoothly Converging and Tunable Free-Energy Method. Phys. Rev. Lett. 2008, 100, 020603.
- Hamelberg, D.; Mongan, J.; McCammon, J.A. Accelerated molecular dynamics: A promising and efficient simulation method for biomolecules. J. Chem. Phys. 2004, 120, 11919–11929.
- Torrie, G.M.; Valleau, J.P. Nonphysical sampling distributions in Monte Carlo free-energy estimation: Umbrella sampling. J. Comput. Phys. 1977, 23, 187–199.
- Kästner, J. Umbrella sampling. WIREs Comput. Mol. Sci. 2011, 1, 932–942.
- Gao, Y.Q. An integrate-over-temperature approach for enhanced sampling. J. Chem. Phys. 2008, 128, 064105.
- MacCallum, J.L.; Muniyat, M.I.; Gaalswyk, K. Online Optimization of Total Acceptance in Hamiltonian Replica Exchange Simulations. J. Phys. Chem. B 2018, 122, 5448–5457.
- Liu, N.; Guo, Y.; Ning, S.; Duan, M. Phosphorylation regulates the binding of intrinsically disordered proteins via a flexible conformation selection mechanism. Commun. Chem. 2020, 3, 123.
- Dickson, A.; Ahlstrom, L.S.; Brooks III, C.L. Coupled folding and binding with 2D Window-Exchange Umbrella Sampling. J. Comput. Chem. 2016, 37, 587–594.
- Sidky, H.; Chen, W.; Ferguson, A.L. Machine learning for collective variable discovery and enhanced sampling in biomolecular simulation. Mol. Phys. 2020, 118, e1737742.
- Chen, W.; Tan, A.R.; Ferguson, A.L. Collective variable discovery and enhanced sampling using autoencoders: Innovations in network architecture and error function design. J. Chem. Phys. 2018, 149, 072312.
- Marinari, E.; Parisi, G. Simulated Tempering: A New Monte Carlo Scheme. EPL (Europhys. Lett.) 1992, 19, 451–458.
- Kamiya, M.; Sugita, Y. Flexible selection of the solute region in replica exchange with solute tempering: Application to protein-folding simulations. J. Chem. Phys. 2018, 149, 072304.
- Wang, L.; Friesner, R.A.; Berne, B.J. Replica Exchange with Solute Scaling: A More Efficient Version of Replica Exchange with Solute Tempering (REST2). J. Phys. Chem. B 2011, 115, 9431–9438.
- Miao, Y.; Sinko, W.; Pierce, L.; Bucher, D.; Walker, R.C.; McCammon, J.A. Improved Reweighting of Accelerated Molecular Dynamics Simulations for Free Energy Calculation. J. Chem. Theory Comput. 2014, 10, 2677–2689.
- Kokubo, H.; Tanaka, T.; Okamoto, Y. Two-dimensional replica-exchange method for predicting protein–ligand binding structures. J. Comput. Chem. 2013, 34, 2601–2614.
- Oshima, H.; Re, S.; Sugita, Y. Replica-Exchange Umbrella Sampling Combined with Gaussian Accelerated Molecular Dynamics for Free-Energy Calculation of Biomolecules. J. Chem. Theory Comput. 2019, 15, 5199–5208.
- Peng, X.; Zhang, Y.; Li, Y.; Liu, Q.; Chu, H.; Zhang, D.; Li, G. Integrating Multiple Accelerated Molecular Dynamics to Improve Accuracy of Free Energy Calculations. J. Chem. Theory Comput. 2018, 14, 1216–1227.
- Bussi, G.; Laio, A. Using metadynamics to explore complex free-energy landscapes. Nat. Rev. Phys. 2020, 2, 200–212.
- Galvelis, R.; Sugita, Y. Replica state exchange metadynamics for improving the convergence of free energy estimates. J. Comput. Chem. 2015, 36, 1446–1455.
- Piana, S.; Laio, A. A Bias-Exchange Approach to Protein Folding. J. Phys. Chem. B 2007, 111, 4553–4559.
- Galvelis, R.; Re, S.; Sugita, Y. Enhanced Conformational Sampling of N-Glycans in Solution with Replica State Exchange Metadynamics. J. Chem. Theory Comput. 2017, 13, 1934–1942.
- Do, T.N.; Choy, W.-Y.; Karttunen, M. Binding of Disordered Peptides to Kelch: Insights from Enhanced Sampling Simulations. J. Chem. Theory Comput. 2016, 12, 395–404.
- Shrestha, U.R.; Smith, J.C.; Petridis, L. Full structural ensembles of intrinsically disordered proteins from unbiased molecular dynamics simulations. Commun. Biol. 2021, 4, 243.
- Hicks, A.; Zhou, H.-X. Temperature-induced collapse of a disordered peptide observed by three sampling methods in molecular dynamics simulations. J. Chem. Phys. 2018, 149, 072313.
- Brown, A.H.; Rodger, P.M.; Evans, J.S.; Walsh, T.R. Equilibrium Conformational Ensemble of the Intrinsically Disordered Peptide n16N: Linking Subdomain Structures and Function in Nacre. Biomacromolecules 2014, 15, 4467–4479.
- Pang, X.; Zhou, H.-X. Disorder-to-Order Transition of an Active-Site Loop Mediates the Allosteric Activation of Sortase A. Biophys. J. 2015, 109, 1706–1715.
- Liu, X.; Jia, Z.; Chen, J. Enhanced Sampling of Intrinsic Structural Heterogeneity of the BH3-Only Protein Binding Interface of Bcl-xL. J. Phys. Chem. B 2017, 121, 9160–9168.
- Liang, C.; Savinov, S.N.; Fejzo, J.; Eyles, S.J.; Chen, J. Modulation of Amyloid-beta42 Conformation by Small Molecules Through Nonspecific Binding. J. Chem. Theory Comput. 2019, 15, 5169–5174.
- Liu, X.; Chen, J. Modulation of p53 Transactivation Domain Conformations by Ligand Binding and Cancer-Associated Mutations. Pac. Symp. Biocomput. 2020, 25, 195–206.
- Schrag, L.G.; Liu, X.; Thevarajan, I.; Prakash, O.; Zolkiewski, M.; Chen, J. Cancer-Associated Mutations Perturb the Disordered Ensemble and Interactions of the Intrinsically Disordered p53 Transactivation Domain. J. Mol. Biol. 2021, 433, 167048.
- Zhao, J.; Blayney, A.; Liu, X.; Gandy, L.; Jin, W.; Yan, L.; Ha, J.H.; Canning, A.J.; Connelly, M.; Yang, C.; et al. EGCG binds intrinsically disordered N-terminal domain of p53 and disrupts p53-MDM2 interaction. Nat. Commun. 2021, 12, 986.
- Wang, L.; Wu, Y.; Deng, Y.; Kim, B.; Pierce, L.; Krilov, G.; Lupyan, D.; Robinson, S.; Dahlgren, M.K.; Greenwood, J.; et al. Accurate and Reliable Prediction of Relative Ligand Binding Potency in Prospective Drug Discovery by Way of a Modern Free-Energy Calculation Protocol and Force Field. J. Am. Chem. Soc. 2015, 137, 2695–2703.
