Genetic engineering and transgenesis provide the opportunity for more significant gains and production in a short span of time. One of the best strategies is the genetic alteration of livestock to enhance the efficiency of food production (e.g., meat and milk), animal health, and welfare (animal population and disease). Moreover, genome engineering in the bovine is majorly focused on subjects such as disease resistance (e.g., tuberculosis), eradicate allergens (e.g., beta-lactoglobulin knock-out), products generation (e.g., meat from male and milk from female), male or female birth specifically (animal sexing), the introduction of valuable traits (e.g., stress tolerance and disease resistance) and their wellbeing (e.g., hornlessness).
- genome editing
- ZFNs
- TALANs
- CRISPR-Cas9
- guide RNA
- livestock
- precision
- specificity
1. Introduction
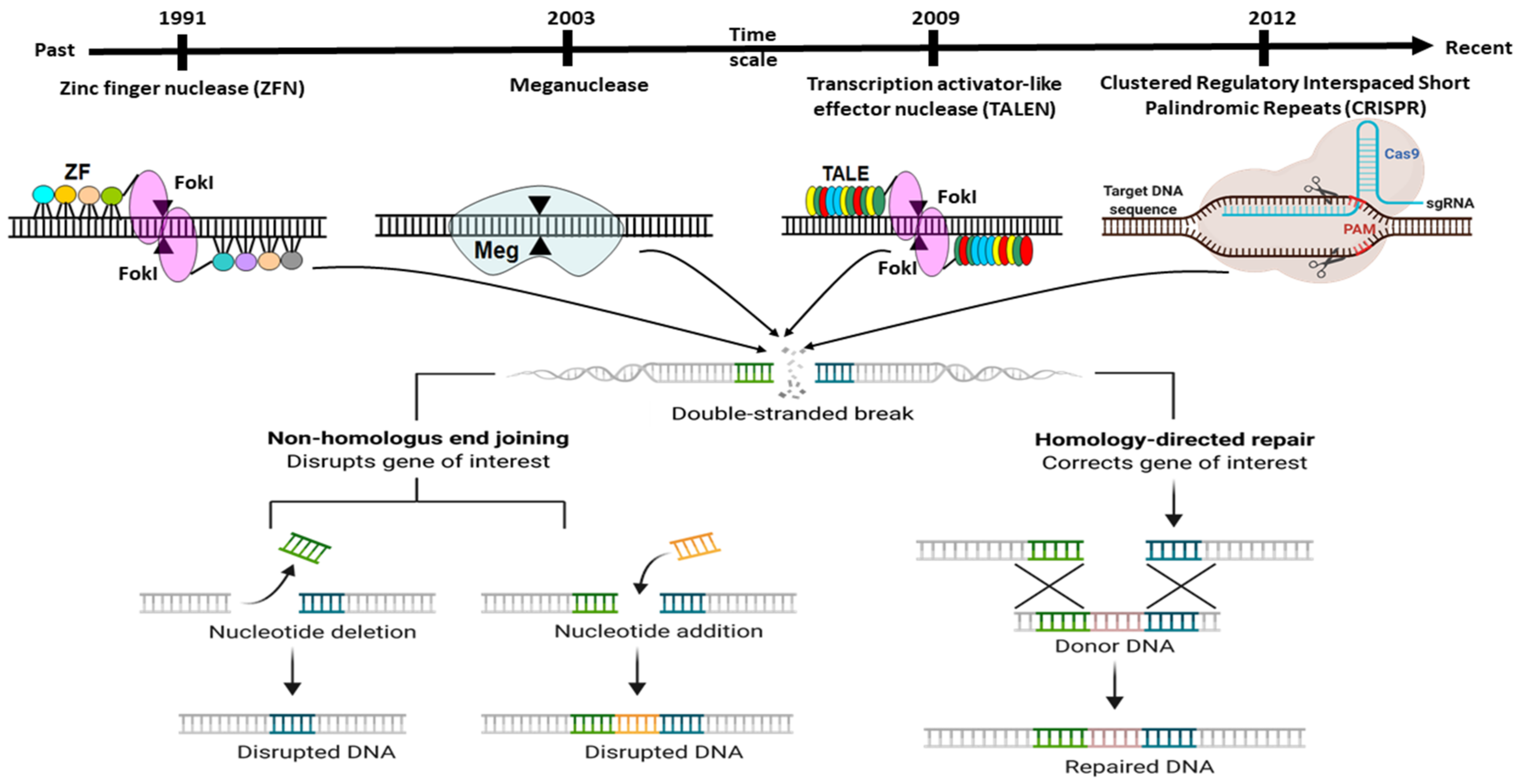
2. Adaptation of Adoptive Mechanism as CRISPR Editing System
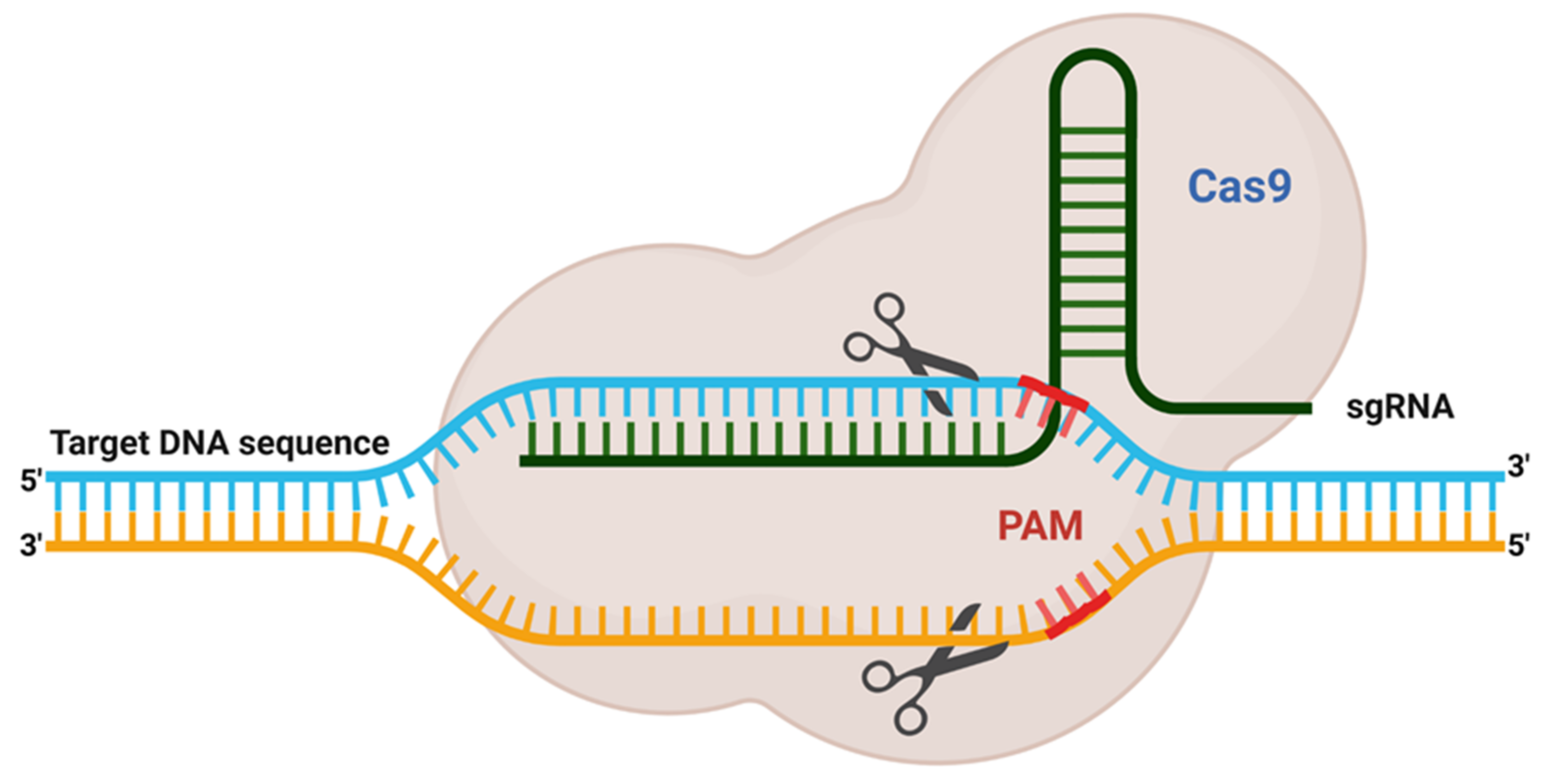
3. Bioinformatics Tool Used to Design sgRNA for Gene Editing
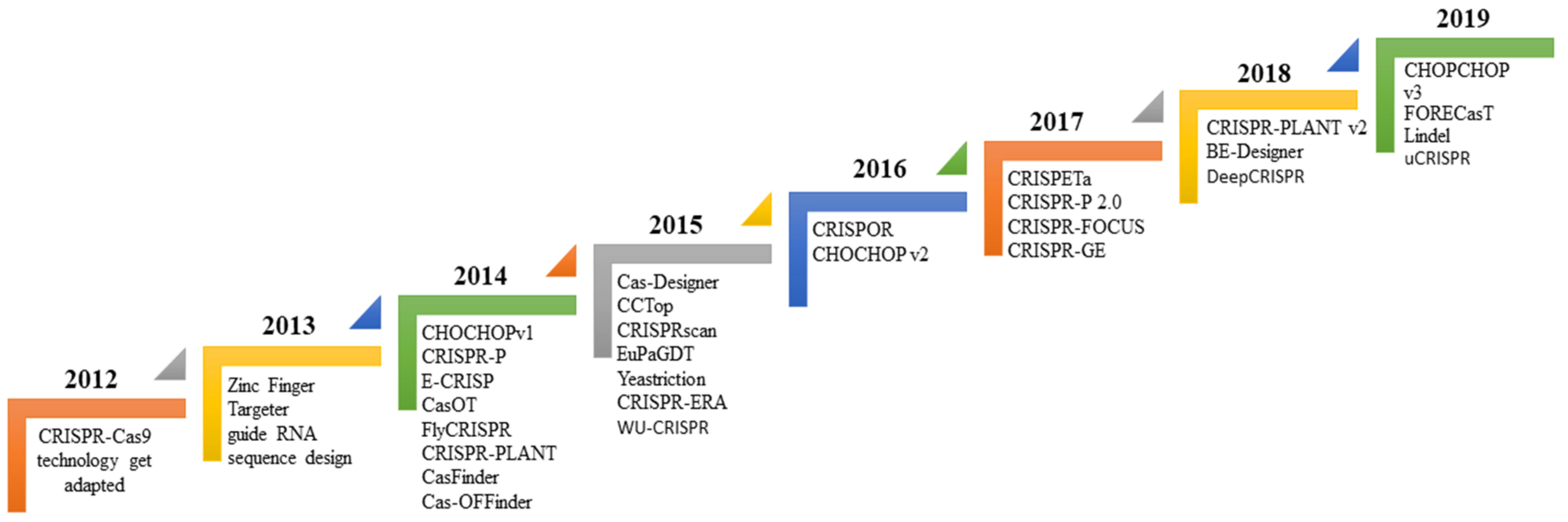
4. Guide RNA Sequence Features
Evaluation Guide RNA Efficiency | Link to Access the Algorithms | ||||
|---|---|---|---|---|---|
E-CRISP (Cas9) | |||||
CRISPRscan (Cas9, Cpf1) | |||||
evaluateCrispr (Cas9) | https://eu.idtdna.com/site/order/designtool/index/CRISPR_SEQUENCE | ||||
sgRNAScorer (Cas9, Cpf1) | |||||
SSC (Cas9) | |||||
WU-CRISPR (Cas9) | |||||
Azimuth (Cas9) | |||||
CRISPRater (Cas9) | |||||
CRISPRpred (Cas9) | https://bmcbioinformatics.biomedcentral.com/articles/10.1186/s12859-020-3531-9 | ||||
CASPER (Cas9, Cpf1) | |||||
DeepCpf1 (Cpf1) | |||||
TSAM (Cas9) | |||||
TUSCAN (Cas9) | |||||
uCRISPR (Cas9) | |||||
Predict guide RNA specificity | |||||
CasOT (Cas9) | |||||
Cas-OFFinder (custom) | |||||
sgRNAcas9 (Cas9) | |||||
FlashFry (custom) | |||||
Crisflash (Cas9) | |||||
MIT (Cas9) | |||||
CCTop (Cas9, Cpf1) | |||||
CFD (Cas9) | https://www.genscript.com/gRNA-detail/mouse/11537/Cas9/Cfd-CRISPR-guide-RNA.html | ||||
CRISPRoff (Cas9) | https://www.genscript.com/gRNA-detail/mouse/11537/Cas9/Cfd-CRISPR-guide-RNA.html | ||||
uCRISPR (Cas9) | |||||
CRISTA (Cas9) | |||||
Elevation (Cas9) | |||||
DeepCRISPR (Cas9) | |||||
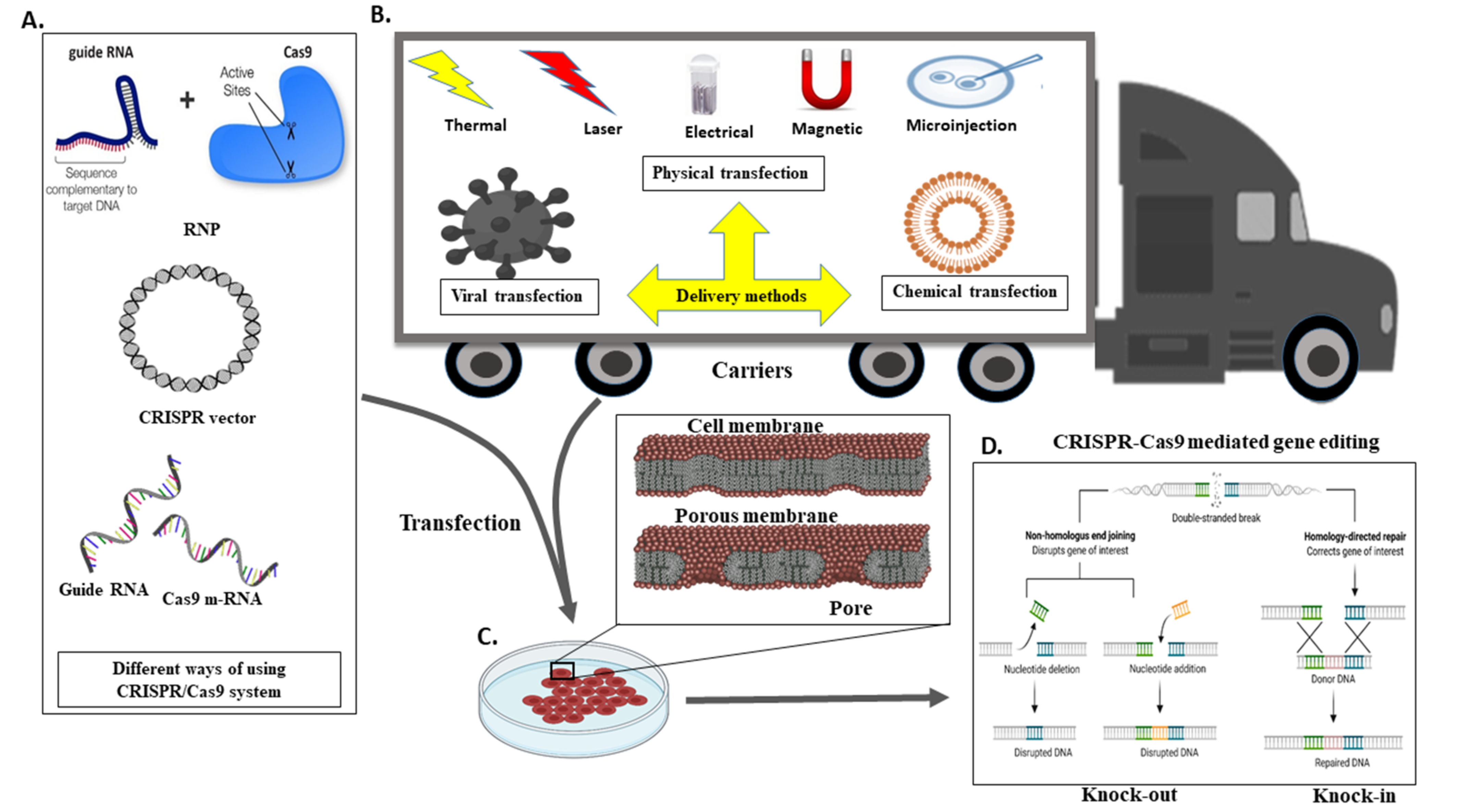
5. A Different Mechanism for the Transport CRISPR System
6. Genome Editing in Ruminants Such as Cattle and Buffalos
It has been anticipated that in the population of 7.6 billion humans globally, every ninth individual (821 million people) does not have sufficient food to cover an active life [81][49]. Despite the lack of food, the human population is expected to rise to 8.5 billion in 2030, 9.7 billion in 2050, and 11.2 billion in 2100 [82][50]. As a result, the United Nations’ Food and Agriculture Organization (FAO) predicts that total agricultural yield (crop yield and animal-based products) should rise to 60% to fulfil global demand. More specifically, this percentage is further contributed to by animal protein, such as meat production by 76%, and milk productivity will need to increase by 63%. In order to achieve this ultimatum goal, a precise and practical approach should be used [84][51]. Meanwhile, genomics targets for genome engineering are possible by screening the differential expression using high throughput proteomics or genomics techniques [85,86,87,88][52][53][54][55]. In this regard, the generation of collective knowledge across the globe allows one to share and build the more efficient farm animals breeds [89][56]. Genome-editing and transgenic innovations offer the chance for more significant gains over a shorter time. Until now, genome editing investigation in cattle has centred fundamentally on enhancing the efficiency of food productivity (e.g., meat and milk), animal health, and welfare (animal population, surveyed or hornlessness and disease), generate all-male offspring, eradication of allergens from products (e.g., beta-lactoglobulin knock-out). On the other hand, genome editing might be utilised to precisely knock-in valuable alleles (such as heat tolerance, illness resistance), as well as haplotypes into our native locally well-adapted cattle breeds genome, subsequently to improve their productivity [90][57]. We recently used the buffalo mammary epithelial cells to understand lactogenic signalling [91,92][58][59]. Early research was majorly focused on animal growth. Skeletal muscle gives meat for human utilisation or consumption, consisting of muscle fibres, intramuscular adipose tissues, and connective tissues [93][60]. The importance of growth hormone (GH) and insulin-like growth factor I (IGF-I) in regulating body size in developing animals has long been recognised. GH and IGF-I play an essential role in muscle growth, both before and after birth [94][61]. The GH–IGF axis (growth hormone-insulin-like growth factor axis), regulated by the pituitary gland and liver, is responsible for muscle growth and body mass [76][62]. GH, on the other hand, induces the development of IGF-I in almost all tissues. The liver is the only organ that can primarily produce serum IGF-I. The pituitary gland produces GH, which stimulates the development of IGF-I in other tissues (liver and muscle). Even though some cIGF-I is released from other tissues, such as muscle, the liver is the most common source of circulating IGF-I (cIGF-I). cIGF-I is a component of the negative feedback loop that controls GH secretion [94][61]. IGF-1 derived from both muscle and liver plays a crucial role in myogenesis [95][63]. At the same time, a mutation in the IGF-2 gene’s regulatory function has been linked to increased muscle growth in pigs [96][64]. In recent years, effective microinjection of the GH and IGF-1 genes into pig zygotes has been reported. Later, two lines of GH-expressing transgene pigs gained 11.1 and 13.7 percent more mass than control pigs [97,98,99][65][66][67]. When transgenic technology is combined with recent genome editing technology, it creates a new age or property for animal protein that could affect animal welfare, while meeting human diet demands. The cloned pig, for example, that expresses the fat-1 gene from the nematode C. elegans, has a lower ratio of n–6 to n–3 fatty acids. A higher ratio of n–6 to n–3 fatty acids has been linked to poor bone health in humans. A lower ratio is related to healthier bone properties; thus, reducing both fatty acids can have nutritional health benefits in a diet [100][68]. Furthermore, related modifications have been observed in pigs containing the C. elegans n–3 fatty acid desaturase gene (encoded by the fat-1 gene) [101,102,103][69][70][71]. Similar findings were obtained when CRISPR/Cas9 was used to insert the fat-1 gene into the pig in the rosa 26 locus [104][72]. This is in proximity with gene alteration (genetic manipulation), which depends on the internalisation of the artificial gene (transgenes) to improve characteristic traits in animals. The genome/gene editing method allows us to make precise and error-free modifications to a livestock animal’s genome, to increase productivity, production, and infection resistance. In the genome editing region, targeted gene editing of the myostatin gene is a popular goal for increasing growth and muscle production. They were first noticed in heavily muscled sheep and cattle like Piedmontese and Belgian Blue cattle and the Texel sheep breed. Additionally, it was discovered that decreased expression of the myostatin gene (also known as GDF8, or growth differentiation factor 8) results in increased muscle growth. Single-nucleotide polymorphisms in the myostatin gene trigger a fundamental genetic change. The Piedmontese and Belgian Blue have a single-nucleotide polymorphism in the myostatin gene and an 11-bp deletion in the myostatin gene [104,105,106][72][73][74].References
- Boch, J. Move over ZFNs. Nat. Biotechnol. 2011, 29, 681–684.
- Cermak, T.; Doyle, E.L.; Christian, M.; Wang, L.; Zhang, Y.; Schmidt, C.; Baller, J.A.; Somia, N.V.; Bogdanove, A.J.; Voytas, D.F. Efficient design and assembly of custom TALEN and other TAL effector-based constructs for DNA targeting. Nucleic Acids Res. 2011, 39, e82.
- Lin, L.; Luo, Y. Tracking CRISPR’s Footprints. Methods Mol. Biol. 2019, 1961, 13–28.
- Barrangou, R.; Fremaux, C.; Deveau, H.; Richards, M.; Boyaval, P.; Moineau, S.; Romero, D.A.; Horvath, P. CRISPR provides acquired resistance against viruses in prokaryotes. Science 2007, 315, 1709–1712.
- Sander, J.D.; Joung, J.K. CRISPR-Cas systems for editing, regulating and targeting genomes. Nat. Biotechnol. 2014, 32, 347–350.
- Sapranauskas, R.; Gasiunas, G.; Fremaux, C.; Barrangou, R.; Horvath, P.; Siksnys, V. The Streptococcus thermophilus CRISPR/Cas system provides immunity in Escherichia coli. Nucleic Acids Res. 2011, 39, 9275–9282.
- Horvath, P.; Barrangou, R. CRISPR/Cas, the immune system of bacteria and archaea. Science 2010, 327, 167–170.
- Makarova, K.S.; Wolf, Y.I.; Iranzo, J.; Shmakov, S.A.; Alkhnbashi, O.S.; Brouns, S.J.; Charpentier, E.; Cheng, D.; Haft, D.H.; Horvath, P.; et al. Evolutionary classification of CRISPR—Cas systems: A burst of class 2 and derived variants. Nat. Rev. Microbiol. 2020, 18, 67–83.
- Fineran, P.C. Resistance is not futile: Bacterial ‘innate’ and CRISPR-Cas ‘adaptive’ immune systems. Microbiology 2019, 165, 834–841.
- Westra, E.R.; Van Houte, S.; Gandon, S.; Whitaker, R. The ecology and evolution of microbial CRISPR-Cas adaptive immune systems. Philos. Trans. R. Soc. B Biol. Sci. 2019, 374, 20190101.
- Barrangou, R.; Marraffini, L.A. CRISPR-Cas systems: Prokaryotes upgrade to adaptive immunity. Mol. Cell 2014, 54, 234–244.
- Kick, L.; Kirchner, M.; Schneider, S. CRISPR-Cas9: From a bacterial immune system to genome-edited human cells in clinical trials. Bioengineered 2017, 8, 280–286.
- Fineran, P.C.; Charpentier, E. Memory of viral infections by CRISPR-Cas adaptive immune systems: Acquisition of new information. Virology 2012, 434, 202–209.
- Zheng, Y.; Li, J.; Wang, B.; Han, J.; Hao, Y.; Wang, S.; Ma, X.; Yang, S.; Ma, L.; Yi, L.; et al. Endogenous type I CRISPR-Cas: From foreign DNA defense to prokaryotic engineering. Front. Bioeng. Biotechnol. 2020, 8, 62.
- Makarova, K.S.; Wolf, Y.I.; Koonin, E.V. Classification and nomenclature of CRISPR-Cas systems: Where from here? CRISPR J. 2018, 1, 325–336.
- Nishimasu, H.; Ran, F.A.; Hsu, P.D.; Konermann, S.; Shehata, S.I.; Dohmae, N.; Ishitani, R.; Zhang, F.; Nureki, O. Crystal structure of Cas9 in complex with guide RNA and target DNA. Cell 2014, 156, 935–949.
- Jinek, M.; Jiang, F.; Taylor, D.W.; Sternberg, S.H.; Kaya, E.; Ma, E.; Anders, C.; Hauer, M.; Zhou, K.; Lin, S. Structures of Cas9 endonucleases reveal RNA-mediated conformational activation. Science 2014, 343, 6176.
- Johnson, R.D.; Jasin, M. Double-strand-break-induced homologous recombination in mammalian cells. Biochem. Soc. Trans. 2001, 29, 196–201.
- Galli, A.; Schiestl, R.H. Effects of DNA double-strand and single-strand breaks on intrachromosomal recombination events in cell-cycle-arrested yeast cells. Genetics 1998, 149, 1235–1250.
- Storici, F.; Durham, C.L.; Gordenin, D.A.; Resnick, M.A. Chromosomal site-specific doublestrand breaks are efficiently targeted for repair by oligonucleotides in yeast. Proc. Natl. Acad. Sci. USA 2003, 100, 14994–14999.
- Lieber, M.R. The mechanism of double-strand DNA break repair by the nonhomologous DNA end-joining pathway. Annu. Rev. Biochem. 2010, 79, 181–211.
- Hsu, P.D.; Scott, D.A.; Weinstein, J.A.; Ran, F.A.; Konermann, S.; Agarwala, V.; Li, Y.; Fine, E.J.; Wu, X.; Shalem, O.; et al. DNA targeting specificity of RNA-guided Cas9 nucleases. Nat. Biotechnol. 2013, 31, 827–832.
- Santiago, Y.; Chan, E.; Liu, P.Q.; Orlando, S.; Zhang, L.; Urnov, F.D.; Holmes, M.C.; Guschin, D.; Waite, A.; Miller, J.C.; et al. Targeted gene knockout in mammalian cells by using engineered zinc-finger nucleases. Proc. Natl. Acad. Sci. USA 2008, 105, 5809–5814.
- Hauschild, J.; Petersen, B.; Santiago, Y.; Queisser, A.L.; Carnwath, J.W.; Lucas-Hahn, A.; Zhang, L.; Meng, X.; Gregory, P.D.; Schwinzer, R.; et al. Efficient generation of a biallelic knockout in pigs using zinc-finger nucleases. Proc. Natl. Acad. Sci. USA 2011, 108, 12013–12017.
- Yang, D.; Yang, H.; Li, W.; Zhao, B.; Ouyang, Z.; Liu, Z.; Zhao, Y.; Fan, N.; Song, J.; Tian, J.; et al. Generation of PPARγ mono-allelic knockout pigs via zinc-finger nucleases and nuclear transfer cloning. Cell Res. 2011, 21, 979–982.
- Yu, S.; Luo, J.; Song, Z.; Ding, F.; Dai, Y.; Li, N. Highly efficient modification of beta-lactoglobulin (BLG) gene via zinc-finger nucleases in cattle. Cell Res. 2011, 21, 1638–1640.
- Liu, X.; Wang, Y.; Guo, W.; Chang, B.; Liu, J.; Guo, Z.; Quan, F.; Zhang, Y. Zinc-finger nickase-mediated insertion of the lysostaphin gene into the beta-casein locus in cloned cows. Nat. Commun. 2013, 4, 2565.
- Epinat, J.C.; Arnould, S.; Chames, P.; Rochaix, P.; Desfontaines, D.; Puzin, C.; Patin, A.; Zanghellini, A.; Pâques, F.; Lacroix, E. A novel engineered meganuclease induces homologous recombination in yeast and mammalian cells. Nucleic Acids Res. 2003, 31, 2952–2962.
- Hockemeyer, D.; Wang, H.; Kiani, S.; Lai, C.S.; Gao, Q.; Cassady, J.P.; Cost, G.J.; Zhang, L.; Santiago, Y.; Miller, J.C. Genetic engineering of human pluripotent cells using TALE nucleases. Nat. Biotechnol. 2011, 29, 731–734.
- Bedell, V.M.; Wang, Y.; Campbell, J.M.; Poshusta, T.L.; Starker, C.G.; Krug, R.G., II; Tan, W.; Penheiter, S.G.; Ma, A.C.; Leung, A.Y.; et al. In vivo genome editing using a high-efficiency TALEN system. Nature 2012, 491, 114–118.
- Carlson, D.F.; Tan, W.; Lillico, S.G.; Stverakova, D.; Proudfoot, C.; Christian, M.; Voytas, D.F.; Long, C.R.; Whitelaw, C.B.A.; Fahrenkrug, S.C. Efficient TALEN-mediated gene knockout in livestock. Proc. Natl. Acad. Sci. USA 2012, 109, 17382–17387.
- Proudfoot, C.; Carlson, D.F.; Huddart, R.; Long, C.R.; Pryor, J.H.; King, T.J.; Lillico, S.G.; Mileham, A.J.; McLaren, D.G.; Whitelaw, C.B.A.; et al. Genome edited sheep and cattle. Transgenic Res. 2015, 24, 147–153.
- Ran, F.A.; Hsu, P.D.; Lin, C.Y.; Gootenberg, J.S.; Konermann, S.; Trevino, A.E.; Scott, D.A.; Inoue, A.; Matoba, S.; Zhang, Y.; et al. Double nicking by RNA-guided CRISPR Cas9 for enhanced genome editing specificity. Cell 2013, 154, 1380–1389.
- Mashiko, D.; Young, S.A.; Muto, M.; Kato, H.; Nozawa, K.; Ogawa, M.; Noda, T.; Kim, Y.J.; Satouh, Y.; Fujihara, Y.; et al. Feasibility for a large scale mouse mutagenesis by injecting CRISPR/Cas plasmid into zygotes. Dev. Growth Differ. 2014, 56, 122–129.
- Pavletich, N.P.; Pabo, C.O. Zinc finger-DNA recognition: Crystal structure of a Zif268-DNA complex at 2.1 A. Science 1991, 252, 809–817.
- Baker, M. Gene-editing nucleases. Nat. Methods 2012, 23–26.
- Jinek, M.; Chylinski, K.; Fonfara, I.; Hauer, M.; Doudna, J.A.; Charpentier, E. A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science 2012, 337, 816–821.
- Deltcheva, E.; Chylinski, K.; Sharma, C.M.; Gonzales, K.; Chao, Y.; Pirzada, Z.A.; Eckert, M.R.; Vogel, J.; Charpentier, E. CRISPR RNA maturation by trans-encoded small RNA and host factor RNase III. Nature 2011, 471, 602–607.
- Wong, N.; Liu, W.; Wang, X. WU-CRISPR: Characteristics of functional guide RNAs for the CRISPR/Cas9 system. Genome Biol. 2015, 16, 218.
- Xu, H.; Xiao, T.F.; Chen, C.H.; Li, W.; Meyer, C.A.; Wu, Q. Sequence determinants of improved CRISPR sgRNA design. Genome Res. 2015, 25, 1147–1157.
- Wu, X.B.; Scott, D.A.; Kriz, A.J.; Chiu, A.C.; Hsu, P.D.; Dadon, D.B. Genome-wide binding of the CRISPR endonuclease Cas9 in mammalian cells. Nat. Biotechnol. 2014, 32, 670–676.
- Liu, G.; Zhang, Y.; Zhang, T. Computational approaches for effective CRISPR guide RNA design and evaluation. Comput. Struct. Biotechnol. J. 2020, 18, 35–44.
- Szczelkun, M.D.; Tikhomirova, M.S.; Sinkunas, T.; Gasiunas, G.; Karvelis, T.; Pschera, P. Direct observation of R-loop formation by single RNA-guided Cas9 and Cascade effector complexes. Proc. Natl. Acad. Sci. USA 2014, 111, 9798–9803.
- Xu, X.; Duan, D.; Chen, S.J. CRISPR-Cas9 cleavage efficiency correlates strongly with target-sgRNA folding stability: From physical mechanism to off-target assessment. Sci. Rep. 2017, 7, 143.
- Jensen, K.T.; Floe, L.; Petersen, T.S.; Huang, J.; Xu, F.; Bolund, L. Chromatin accessibility and guide sequence secondary structure affect CRISPR-Cas9 gene editing efficiency. FEBS Lett. 2017, 591, 1892–1901.
- Doench, J.G.; Fusi, N.; Sullender, M.; Hegde, M.; Vaimberg, E.W.; Donovan, K.F. Optimized sgRNA design to maximise activity and minimise off-target effects of CRISPR-Cas9. Nat. Biotechnol. 2016, 34, 184–191.
- Lino, C.A.; Harper, J.C.; Carney, J.P.; Timlin, J.A. Delivering CRISPR: A review of the challenges and approaches. Drug Deliv. 2018, 25, 1234–1257.
- Mittal, D.; Kaur, G.; Singh, P.; Yadav, K.; Ali, S.A. Nanoparticle-based sustainable agriculture and food science: Recent advances and future outlook. Front. Nanotechnol. 2020, 2, 10.
- Hunger Map; World Food Program; United Nations World Food Programme—Fighting Hunger Worldwide. Available online: https://www.wfp.org/content/2018-hunger-map (accessed on 30 October 2018).
- United Nations. World Population Prospects. 2015. Available online: https://population.un.org/wpp/Publications/Files/Key_Findings_WPP_2015.pdf (accessed on 30 October 2018).
- Alexandratos, N.; Bruinsma, J. World Agriculture towards 2030/2050: The 2012 Revision 2030. Available online: http://www.fao.org/fileadmin/templates/esa/Global (accessed on 7 June 2021).
- Chopra, A.; Ali, S.A.; Bathla, S.; Rawat, P.; Vohra, V.; Kumar, S.; Mohanty, A.K. High-Resolution Mass Spectrometer—Based Ultra-Deep Profile of Milk Whey Proteome in Indian Zebu (Sahiwal) Cattle. Front. Nutr. 2020, 7, 150.
- Lotfan, M.; Ali, S.A.; Yadav, M.L.; Choudhary, S.; Jena, M.K.; Kumar, S.; Mohanty, A.K. Genome-wide gene expression analysis of 45 days pregnant fetal cotyledons vis-a-vis non-pregnant caruncles in buffalo (Bubalus bubalis). Gene 2018, 654, 127–137.
- Shashikumar, N.G.; Baithalu, R.K.; Bathla, S.; Ali, S.A.; Rawat, P.; Kumaresan, A.; Kumar, S.; Maharana, B.R.; Singh, G.; Kumar, D.P.; et al. Global proteomic analysis of water buffalo (Bubalus bubalis) saliva at different stages of estrous cycle using high throughput mass spectrometry. Theriogenology 2018, 110, 52–60.
- Rawat, P.; Bathla, S.; Baithalu, R.; Yadav, M.L.; Kumar, S.; Ali, S.A.; Tiwari, A.; Lotfan, M.; Naru, J.; Jena, M.; et al. Identification of potential protein biomarkers for early detection of pregnancy in cow urine using 2D DIGE and label free quantitation. Clin. Proteom. 2016, 13, 1–14.
- Almeida, A.M.; Ali, S.A.; Ceciliani, F.; Eckersall, P.D.; Hernandez-Castellano, L.E.; Han, R.; Hodnik, J.J.; Jaswal, S.; Lippolis, J.D.; McLaughlin, M.; et al. Domestic animal proteomics in the 21st century: A global retrospective and viewpoint analysis. J. Proteom. 2021, 241, 104220.
- Van Eenennaam, A.L. Application of genome editing in farm animals: Cattle. In Transgenic Research; Springer International: New York, NY, USA, 2019; Volume 28, pp. 93–100.
- Verma, A.K.; Ali, S.A.; Singh, P.; Kumar, S.; Mohanty, A.K. Transcriptional repression of MFG-E8 causes disturbance in the homeostasis of Cell cycle through DOCK/ZP4/STAT signaling in buffalo mammary epithelial cells. Front. Cell Dev. Biol. 2021, 9, 568660.
- Jaswal, S.; Anand, V.; Ali, S.A.; Jena, M.K.; Kumar, S.; Kaushik, J.K.; Mohanty, A.K. TMT based deep proteome analysis of buffalo mammary epithelial cells and identification of novel protein signatures during lactogenic differentiation. FASEB J. 2021, 35, e21621.
- Zhao, L.; Huang, Y.; Du, M. Farm animals for studying muscle development and metabolism: Dual purposes for animal production and human health. Anim. Front. 2019, 9, 21–27.
- Velloso, C.P. Regulation of muscle mass by growth hormone and IGF-I. Br. J. Pharmacol. 2008, 154, 557–568.
- Liu, H.; Wei, Z.; Dominguez, A.; Li, Y.; Wang, X.; Qi, L.S. CRISPR-ERA: A comprehensive design tool for CRISPR-mediated gene editing, repression and activation. Bioinformatics 2015, 31, 3676–3678.
- Florini, J.R.; Ewton, D.Z.; Coolican, S.A. Growth hormone and the insulin-like growth factor system in myogenesis. Endocr. Rev. 1996, 17, 481–517.
- Van Laere, A.S.; Nguyen, M.; Braunschweig, M.; Nezer, C.; Collette, C.; Moreau, L.; Archibald, A.L.; Haley, C.S.; Buys, N.; Tally, M.; et al. A regulatory mutation in IGF2 causes a major QTL effect on muscle growth in the pig. Nature 2003, 425, 832–836.
- Pursel, V.G.; Bolt, D.J.; Miller, K.F.; Pinkert, C.A.; Hammer, R.E.; Palmiter, R.D.; Brinster, R.L. Expression of Growth Hormone Transgenes in Swine. J. Reprod. Fertil. 1990, 40, 235–245.
- Pursel, V.G.; Wall, R.J.; Mitchell, A.D.; Elsasser, T.H.; Solomon, M.B.; Coleman, M.E.; DeMayo, F.; Schwartz, R.J. Expression of insulin-like growth factor-I in skeletal muscle of transgenic swine. In Transgenic Animals in Agriculture; U.S. Department of Agriculture: Washington, DC, USA, 1999.
- Pursel, V.G.; Pinkert, C.A.; Miller, K.F.; Bolt, D.J.; Campbell, R.G.; Palmiter, R.D.; Brinster, R.L.; Hammer, R.E. Genetic engineering of livestock. Science 1989, 244, 1281–1288.
- Lai, L.; Kang, J.X.; Li, R.; Wang, J.; Witt, W.T.; Yong, H.Y.; Hao, Y.; Wax, D.M.; Murphy, C.N.; Rieke, A.; et al. Generation of cloned transgenic pigs rich in omega-3 fatty acids. Nat. Biotechnol. 2006, 24, 435–436.
- Fiester, A. Why the omega-3 piggy should not go to market. Nat. Biotechnol. 2006, 24, 1472–1473.
- Zhang, P.; Zhang, Y.; Dou, H.; Yin, J.; Chen, Y.; Pang, X.; Vajta, G.; Bolund, L.; Du, Y.; Ma, R.Z. Handmade cloned transgenic piglets expressing the nematode fat-1 gene. Cell. Reprogram. 2012, 14, 258–266.
- Liu, X.; Pang, D.; Yuan, T.; Li, Z.; Li, Z.; Zhang, M.; Ren, W.; Ouyang, H.; Tang, X. N-3 polyunsaturated fatty ac-ids attenuates triglyceride and inflammatory factors level in h fat-1 transgenic pigs. Lipids Health Dis. 2016, 15, 89.
- Grobet, L.; Martin, L.J.R.; Poncelet, D.; Pirottin, D.; Brouwers, B.; Riquet, J.; Schoeberlein, A.; Dunner, S.; Ménis-sier, F.; Massabanda, J.; et al. A deletion in the bovine myostatin gene causes the double–muscled phenotype in cattle. Nat. Genet. 1997, 17, 71–74.
- Kambadur, R.; Sharma, M.; Smith, T.P.; Bass, J.J. Mutations in myostatin (GDF8) in double-muscled Belgian Blue and Piedmontese cattle. Genome Res. 1997, 7, 910–915.
- Tait-Burkard, C.; Doeschl-Wilson, A.; McGrew, M.J.; Archibald, A.L.; Sang, H.M.; Houston, R.D.; Whitelaw, C.B.; Watson, M. Livestock 2.0—Genome editing for fitter, healthier, and more productive farmed animals. Genome Biol. 2018, 19, 204.
