Globally, non-small cell lung cancer (NSCLC) is the leading cause of cancer deaths. The 5-year survival rate has remained at 16% for the past forty years despite advancements in chemotherapy and targeted therapies. Minimal residual disease (MRD) is described as the existence of either isolated tumour cells or circulating tumour cells in biological liquid of patients after removal of the primary tumour without any clinical signs of cancer. Recently, liquid biopsy has been promising as a non-invasive method of disease monitoring and treatment guidelines as an MRD marker. Liquid biopsy could be used to detect and assessment of earlier stages of NSCLC, post-treatment MRD, resistance to targeted therapies, immune checkpoint inhibitors (ICIs) and tumour mutational burden. MRD surveillance has been proposed as a potential marker for lung cancer relapse. Principally biosensors provide the quantitative analysis of various materials by converting biological functions into quantifiable signals. Biosensors are usually operated to detect antibodies, enzymes, DNA, RNA, EVs, and whole cells. Here we present a category of biosensors based on the signal transduction method for identifying biosensor-based biomarkers in liquid biopsy specimens to monitor lung cancer treatment
- bio-sensor
- liquid biopsy
1. Liquid Biopsy as an Alternative Technique for Tissue Biopsy
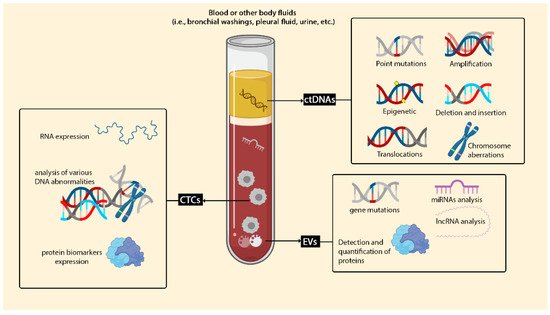
1.1. Circulating Cell-Free DNA (cfDNA) and Circulating Tumour DNA (ctDNA)
1.2. Circulating Tumor Cells (CTCs)
1.3. Extracellular Vesicles (EVs)
2. Type of Biosensors for Detection of Lung Cancer Biomarkers in Liquid Biopsy
| Transduce | Principle | Advantages | Disadvantages | Application in Lung Cancer MRD | ||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Electrochemical sensor | Convert the biochemical interaction to electrical signals |
|
|
| ||||||
| Magnetic biosensor | Applying paramagnetic particles to detect biological interactions by monitoring magnetic property changes |
|
|
| ||||||
| Surface-enhanced Raman spectroscopy (SERS) | Using molecules adsorbing on rough metal surfaces to generate Raman scattering |
|
|
| ||||||
| Surface plasmon resonance biosensors (SPR) | Alteration of SPR angle due to increasing of refractive index during binding of biomolecules on the sensor surface |
|
|
| ||||||
| Fluorescence-based biosensing | An analytical signal of a photoluminescence emission mechanism |
|
|
| ||||||
| Lateral flow immunoassay (LFIA) | Optical properties |
|
|
| ||||||
| Chemiluminescence | Electrochemistry and visual luminescence measurements |
|
|
|
2.1. Electrochemical Sensor
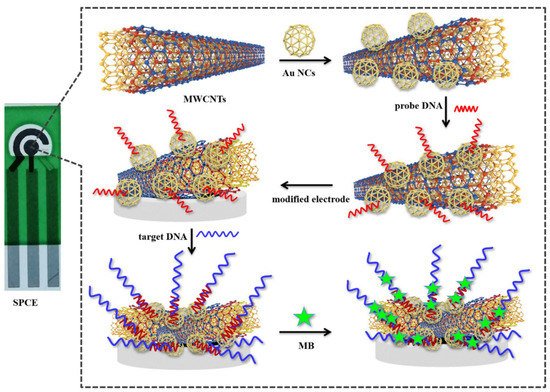
2.2. Electro-Kinetic Biosensors
2.3. Magnetic Biosensor
2.4. Optical Biosensors
2.4.1. Surface Enhanced Raman Spectroscopy (SERS)
2.4.2. Surface Plasmon Resonance Biosensors (SPR)
References
- Cirmena, G.; Garuti, A.; De Mariano, M.; Coco, S.; Ferrando, L.; Isnaldi, E.; Barbero, V.; Fregatti, P.; Del Mastro, L.; Ferrando, F.; et al. Circulating Tumor DNA Using Tagged Targeted Deep Sequencing to Assess Minimal Residual Disease in Breast Cancer Patients Undergoing Neoadjuvant Chemotherapy. J. Oncol. 2020, 2020, 8132507.
- Dizdar, L.; Fluegen, G.; van Dalum, G.; Honisch, E.; Neves, R.P.; Niederacher, D.; Neubauer, H.; Fehm, T.; Rehders, A.; Krieg, A.; et al. Detection of Circulating Tumor Cells in Colorectal Cancer Patients Using the GILUPI CellCollector: Results from a Prospective, Single-Center Study. Mol. Oncol. 2019, 13, 1548–1558.
- Chen, E.; Cario, C.L.; Leong, L.; Lopez, K.; Márquez, C.P.; Chu, C.; Li, P.S.; Oropeza, E.; Tenggara, I.; Cowan, J.; et al. Cell-free DNA Concentration and Fragment Size as a Biomarker for Prostate Cancer. Sci. Rep. 2021, 11, 5040.
- Cheng, F.; Su, L.; Qian, C. Circulating Tumor DNA: A Promising Biomarker in the Liquid Biopsy of Cancer. Oncotarget 2015, 7, 48832–48841.
- Russano, M.; Napolitano, A.; Ribelli, G.; Iuliani, M.; Simonetti, S.; Citarella, F.; Pantano, F.; Dell’Aquila, E.; Anesi, C.; Silvestris, N.; et al. Liquid Biopsy and Tumor Heterogeneity in Metastatic Solid Tumors: The Potentiality of Blood Samples. J. Exp. Clin. Cancer Res. 2020, 39, 95.
- Cucchiara, F.; Del Re, M.; Valleggi, S.; Romei, C.; Petrini, I.; Lucchesi, M.; Crucitta, S.; Rofi, E.; De Liperi, A.; Chella, A.; et al. Integrating Liquid Biopsy and Radiomics to Monitor Clonal Heterogeneity of EGFR-Positive Non-Small Cell Lung Cancer. Front. Oncol. 2020, 10, 2664.
- Peng, M.; Huang, Q.; Yin, W.; Tan, S.; Chen, C.; Liu, W.; Tang, J.; Wang, X.; Zhang, B.; Zou, M.; et al. Circulating Tumor DNA as a Prognostic Biomarker in Localized Non-small Cell Lung Cancer. Front. Oncol. 2020, 10, 561598.
- Abbosh, C.; Birkbak, N.J.; Wilson, G.A.; Jamal-Hanjani, M.; Constantin, T.; Salari, R.; Le Quesne, J.; Moore, D.A.; Veeriah, S.; Rosenthal, R.; et al. Phylogenetic Ctdna Analysis Depicts Early-Stage Lung Cancer Evolution. Nature 2017, 545, 446–451.
- Chen, L.; Bode, A.M.; Dong, Z. Circulating Tumor Cells: Moving Biological Insights into Detection. Theranostics 2017, 7, 2606–2619.
- Tamminga, M.; de Wit, S.; Hiltermann, T.J.N.; Timens, W.; Schuuring, E.; Terstappen, L.; Groen, H.J. Circulating Tumor Cells in Advanced Non-Small Cell Lung Cancer Patients are Associated with Worse Tumor Response to Checkpoint Inhibitors. J. Immunother. 2019, 7, 173.
- Cheng, Y.; Wang, T.; Lv, X.; Li, R.; Yuan, L.; Shen, J.; Li, Y.; Yan, T.; Liu, B.; Wang, L. Detection of PD-L1 Expression and Its Clinical Significance in Circulating Tumor Cells from Patients with Non-Small-Cell Lung Cancer. Cancer Manag. Res. 2020, 12, 2069–2078.
- Kong, D.; Zhang, W.; Yang, Z.; Li, G.; Cheng, S.; Zhang, K.; Feng, L. Correlation between PD-L1 Expression ON CTCs and Prognosis of Patients with Cancer: A Systematic Review and Meta-Analysis. OncoImmunology 2021, 10, 1938476.
- Wu, C.-Y.; Lee, C.-L.; Fu, J.-Y.; Yang, C.-T.; Wen, C.-T.; Liu, Y.-H.; Liu, H.-P.; Hsieh, J.C.-H.; Wu, C.-F. Circulating Tumor Cells as a Tool of Minimal Residual Disease Can Predict Lung Cancer Recurrence: A Longitudinal, Prospective Trial. Diagnostics 2020, 10, 144.
- Li, M.-Y.; Liu, L.-Z.; Dong, M. Progress on Pivotal Role and Application of Exosome in Lung Cancer Carcinogenesis, Diagnosis, Therapy and Prognosis. Mol. Cancer 2021, 20, 22.
- Su, H.; Li, S.; Jin, Y.; Xian, Z.; Yang, D.; Zhou, W.; Mangaran, F.; Leung, F.; Sithamparanathan, G.; Kerman, K. Nanomaterial-Based Biosensors for Biological Detections. Adv. Heal. Care Technol. 2017, 3, 19–29.
- Aykaç, A.; Gergeroglu, H.; Beşli, B.; Akkaş, E.; Yavaş, A.; Güler, S.; Güneş, F.; Erol, M. An Overview on Recent Progress of Metal Oxide/Graphene/CNTs-Based Nanobiosensors. Nanoscale Res. Lett. 2021, 16, 65.
- Naresh, V.; Lee, N. A Review on Biosensors and Recent Development of Nanostructured Materials-Enabled Biosensors. Sensors 2021, 21, 1109.
- Yeh, Y.-C.; Creran, B.; Rotello, V.M. Gold nanoparticles: Preparation, Properties, and Applications in Bionanotechnology. Nanoscale 2011, 4, 1871–1880.
- Tîlmaciu, C.-M.; Morris, M.C. Carbon Nanotube Biosensors. Front. Chem. 2015, 3, 59.
- Chen, M.; Wu, D.; Tu, S.; Yang, C.; Chen, D.; Xu, Y. A Novel Biosensor for the Ultrasensitive Detection of the Lncrna Biomarker MALAT1 in Non-Small Cell Lung Cancer. Sci. Rep. 2021, 11, 3666.
- Muley, T.; He, Y.; Rolny, V.; Wehnl, B.; Escherich, A.; Warth, A.; Stolp, C.; Schneider, M.A.; Meister, M.; Herth, F.J.; et al. Potential for the Blood-Based Biomarkers Cytokeratin 19 Fragment (CYFRA 21-1) and Human Epididymal Protein 4 (HE4) to Detect Recurrence during Monitoring after Surgical Resection of Adenocarcinoma of the Lung. Lung Cancer 2019, 130, 194–200.
- Yola, M.L.; Atar, N.; Özcan, N. A Novel Electrochemical Lung Cancer Biomarker Cytokeratin 19 Fragment Antigen 21-1 Immunosensor Based on Si3N4/Mos2 Incorporated Mwcnts and Core–Shell Type Magnetic Nanoparticles. Nanoscale 2021, 13, 4660–4669.
- Dall’Olio, F.G.; Abbati, F.; Facchinetti, F.; Massucci, M.; Melotti, B.; Squadrilli, A.; Buti, S.; Formica, F.; Tiseo, M.; Ardizzoni, A. CEA and CYFRA 21-1 as Prognostic Biomarker and as a Tool for Treatment Monitoring in Advanced NSCLC Treated with Immune Checkpoint Inhibitors. Adv. Med. Oncol. 2020, 12, 1–13.
- Yoon, H.J.; Kim, T.H.; Zhang, Z.; Azizi, E.; Pham, T.M.; Paoletti, C.; Lin, J.; Ramnath, N.; Wicha, M.S.; Hayes, D.F.; et al. Sensitive Capture of Circulating Tumour Cells by Functionalized Graphene Oxide Nanosheets. Nat. Nanotechnol. 2013, 8, 735–741.
- Bolat, G.; Vural, O.A.; Yaman, Y.T.; Abaci, S. Polydopamine Nanoparticles-Assisted Impedimetric Sensor Towards Label-Free Lung Cancer Cell Detection. Mater. Sci. Eng. C 2021, 119, 111549.
- Nguyen, N.-V.; Jen, C.-P. Selective Detection of Human Lung Adenocarcinoma Cells Based on the Aptamer-Conjugated Self-Assembled Monolayer of Gold Nanoparticles. Micromachines 2019, 10, 195.
- Wei, F.; Lin, C.-C.; Joon, A.; Feng, Z.; Troche, G.; Lira, M.E.; Chia, D.; Mao, M.; Ho, C.L.; Su, W.C.; et al. Noninvasive Saliva-Based EGFR Gene Mutation Detection in Patients with Lung Cancer. Am. J. Respir. Crit. Care Med. 2014, 15, 1117–1126.
- Li, F.; Wei, F.; Huang, W.-L.; Lin, C.-C.; Li, L.; Shen, M.M.; Yan, Q.; Liao, W.; Chia, W.; Tu, M.; et al. Ultra-Short Circulating Tumor DNA (usctDNA) in Plasma and Saliva of Non-Small Cell Lung Cancer (NSCLC) Patients. Cancers 2020, 12, 2041.
- Li, N.; Guha, U.; Kim, C.; Ye, L.; Cheng, J.; Li, F.; Chia, D.; Wei, F.; Wong, D.T.W. Longitudinal Monitoring of EGFR and PIK3CA Mutations by Saliva-Based EFIRM in Advanced NSCLC Patients with Local Ablative Therapy and Osimertinib Treatment: Two Case Reports. Front. Oncol. 2020, 10, 1240.
- Wei, F.; Yang, J.; Wong, D.T.W. Detection of Exosomal Biomarker by Electric Field-Induced Release and Measurement (EFIRM). Biosens. Bioelectron. 2013, 44, 115–121.
- Lindeman, L.R.; Jones, K.M.; High, R.A.; Howison, C.M.; Shubitz, L.F.; Pagel, M.D. Differentiating Lung Cancer and Infection Based on Measurements of Extracellular pH with acidoCEST MRI. Sci. Rep. 2019, 9, 13002.
- Mani, G.K.; Morohoshi, M.; Yasoda, Y.; Yokoyama, S.; Kimura, H.; Tsuchiya, K. ZnO-Based Microfluidic pH Sensor: A Versatile Approach for Quick Recognition of Circulating Tumor Cells in Blood. ACS Appl. Mater. 2017, 9, 5193–5203.
- Ronkainen, N.J.; Halsall, H.B.; Heineman, W.R. Electrochemical Biosensors. Chem. Soc. Rev. 2010, 39, 1747–1763.
- Cavallaro, S.; Hååg, P.; Sahu, S.S.; Berisha, L.; Kaminskyy, V.O.; Ekman, S.; Lewensohn, R.; Linnros, J.; Viktorsson, K.; Dev, A. Multiplexed Electrokinetic Sensor for Detection and Therapy Monitoring of Extracellular Vesicles from Liquid Biopsies of Non-Small-Cell Lung Cancer Patients. Biosens. Bioelectron. 2021, 193, 113568.
- Sahu, S.S.; Stiller, C.; Cavallaro, S.; Karlström, A.E.; Linnros, J.; Dev, A. Influence of Molecular Size and Zeta Potential in Electrokinetic Biosensing. Biosens. Bioelectron. 2020, 152, 11200.
- Feng, J.; Wu, T.; Cheng, Q.; Ma, H.; Ren, X.; Wang, X.; Lee, J.Y.; Wei, Q.; Ju, H. A Microfluidic Cathodic Photoelectrochemical Biosensor Chip for the Targeted Detection of Cytokeratin 19 Fragments 21-1. Lab Chip 2021, 21, 378–384.
- Nabaei, V.; Chandrawati, R.; Heidari, H. Magnetic Biosensors: Modelling and Simulation. Biosens. Bioelectron. 2018, 103, 69–86.
- Zhang, Y.; Yang, D.; Weng, L.; Wang, L. Early Lung Cancer Diagnosis by Biosensors. Int. J. Mol. Sci. 2013, 14, 15479–15509.
- Nair, V.S.; Beggs, M.; Yu, H.; Carbonell, L.; Wang, S.X.; Vachani, A. Validation of Plasma TIMP-1 to Identify Lung Cancer in Smokers. In D99 Clinically Informative Biomarkers in Lung Cancer: A Needle in a Haystack; American Thoracic Society: New York, NY, USA, 2018; p. A7415.
- Pesta, M.; Kulda, V.; Kucera, R.; Pesek, M.; Vrzalova, J.; Liska, V.; Pecen, L.; Treska, V.; Safranek, J.; Prazakova, M.; et al. Prognostic Significance of TIMP-1 in Non-Small Cell Lung Cancer. Anticancer. Res. 2011, 31, 4031–4038.
- Selvaraj, G.; Kaliamurthi, S.; Lin, S.; Gu, K.; Wei, D. Prognostic Impact of Tissue Inhibitor of Metalloproteinase-1 in Non- Small Cell Lung Cancer: Systematic Review and Meta-Analysis. Curr. Med. Chem. 2019, 26, 7694–7713.
- Chang, Y.-H.; Chiu, Y.-J.; Cheng, H.-C.; Liu, F.-J.; Lai, W.-W.; Chang, H.-J.; Liao, P.-C. Down-Regulation of TIMP-1 Inhibits Cell Migration, Invasion, and Metastatic Colonization in Lung Adenocarcinoma. Tumor Biol. 2015, 36, 3957–3967.
- Damborský, P.; Švitel, J.; Katrlík, J. Optical Biosensors. Essays Biochem. 2016, 60, 91–100.
- Li, P.; Long, F.; Chen, W.; Chen, J.; Chu, P.K.; Wang, H. Fundamentals and Applications of Surface-Enhanced Raman Spectroscopy–Based Biosensors. Curr. Opin. Biomed. Eng. 2019, 13, 51–59.
- Pang, Y.; Shi, J.; Yang, X.; Wang, C.; Sun, Z.; Xiao, R. Personalized detection of circling exosomal PD-L1 based on Fe3O4@TiO2 isolation and SERS immunoassay. Biosens. Bioelectron. 2020, 148, 111800.
- Lin, J.; Zheng, J.; Wu, A. An Efficient Strategy for Circulating Tumor Cell Detection: Surface-Enhanced Raman Spectroscopy. J. Mater. Chem. B 2020, 8, 3316–3326.
- Song, C.Y.; Yang, Y.J.; Yang, B.Y.; Sun, Y.Z.; Zhao, Y.P.; Wang, L.H. An Ultrasensitive SERS Sensor for Simultaneous Detection of Multiple Cancer-Related miRNAs. Nanoscale 2016, 8, 17365–17373.
- Fan, Y.; Duan, X.; Zhao, M.; Wei, X.; Wu, J.; Chen, W.; Liu, P.; Cheng, W.; Cheng, Q.; Ding, S. High-Sensitive and Multiplex Biosensing Assay of NSCLC-Derived Exosomes via Different Recognition Sites Based on Spri Array. Biosens. Bioelectron. 2020, 154, 112066.
- Shin, S.; Park, Y.H.; Jung, S.-H.; Jang, S.-H.; Kim, M.Y.; Lee, J.Y.; Chung, Y. Urinary Exosome Microrna Signatures as a Noninvasive Prognostic Biomarker for Prostate Cancer. npj Genom. Med. 2021, 6, 45.
- Hornick, N.I.; Huan, J.; Doron, B.; Goloviznina, N.; Lapidus, J.; Chang, B.; Kurre, P. Serum Exosome MicroRNA as a Minimally-Invasive Early Biomarker of AML. Sci. Rep. 2015, 5, 11295.
- Foster, M.C.; Fedoriw, Y.; Pulley, W.; Zeidner, J.; Coombs, C.C.; Mirkin, E.; Zomorrodi, M.; Toughiri, R.; Bartakova, A.; Carson, C.; et al. Detection of Measurable Residual Disease (MRD) in Peripheral Blood: First Report of a Novel Microfluidic Platform in Patients with Acute Myeloid Leukemia (AML). Blood 2019, 134, 1417.
- Munagala, R.; Aqil, F.; Gupta, R.C. Exosomal miRNAs as Biomarkers of Recurrent Lung Cancer. Tumor Biol. 2016, 37, 10703–10714.
- Wu, J.; Shen, Z. Exosomal miRNAs as Biomarkers for Diagnostic and Prognostic in Lung Cancer. Cancer Med. 2020, 9, 6909–6922.
- Wu, W.; Yu, X.; Wu, J.; Wu, T.; Fan, Y.; Chen, W.; Zhao, M.; Wu, H.; Li, X.; Ding, S. Surface Plasmon Resonance Imaging-Based Biosensor for Multiplex and Ultrasensitive Detection of NSCLC-Associated Exosomal Mirnas Using DNA Programmed Heterostructure of Au-On-Ag. Biosens. Bioelectron. 2021, 175, 112835.
- Liu, C.; Zeng, X.; An, Z.; Yang, Y.; Eisenbaum, M.; Gu, X.; Jornet, J.M.; Dy, G.K.; Reid, M.E.; Gan, Q.; et al. Sensitive Detection of Exosomal Proteins via a Compact Surface Plasmon Resonance Biosensor for Cancer Diagnosis. ACS Sens. 2018, 3, 1471–1479.
