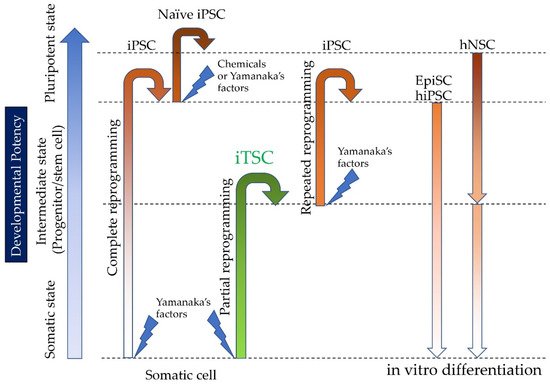
Induced tissue-specific stem cells (iTSCs) are partially reprogrammed cells which have an intermediate state, such as progenitors or stem cells. They originate from the de-differentiation of differentiated somatic cells into pluripotent stem cells, such as induced pluripotent stem cells (iPSCs) and embryonic stem cells (ESCs), or from the differentiation of undifferentiated cells. They show a limited capacity to differentiate and a morphology similar to that of somatic cell stem cells present in tissues, but distinct from that of iPSCs and ESCs. iTSCs can be generally obtained 7 to 10 days after reprogramming of somatic cells with Yamanaka’s factors, and their fibroblast-like morphology remains unaltered. iTSCs can also be obtained directly from iPSCs cultured under conditions allowing cellular differentiation. In this case, to effectively induce iTSCs, additional treatment is required, as exemplified by the conversion of iPSCs into naïve iPSCs. iTSCs can proliferate continuously in vitro, but when transplanted into immunocompromised mice, they fail to generate solid tumors (teratomas), implying loss of tumorigenic potential. The low tendency of iTSCs to elicit tumors is beneficial, especially considering applications for regenerative medicine in humans. Several iTSC types have been identified, including iTS-L, iTS-P, and iTS-D, obtained by reprogramming hepatocytes, pancreatic cells, and deciduous tooth-derived dental pulp cells, respectively. This review provides a brief overview of iPSCs and discusses recent advances in the establishment of iTSCs and their possible applications in regenerative medicine.
- induced tissue-specific stem cells (iTSCs)
- induced pluripotent stem cells (iPSCs)
- partial reprogramming
- naïve stem cells
- epiblast stem cells
- insulin-producing cells
1. Introduction
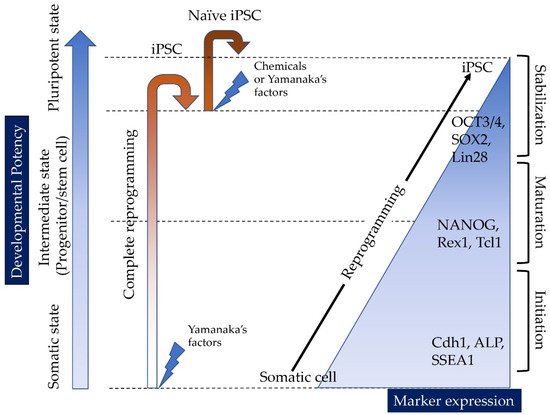
1.1. Reprogramming of Somatic Cells into iPSCs
1.2. Epigenetic Memory
1.3. Potential of Human Embryonic Stem Cells (hESCs)/Human iPSCs (hiPSCs) for Cell-Based Therapies
2. The Discovery of Induced Tissue-Specific Stem Cells (iTSCs), as an Alternative to ESCs/iPCs
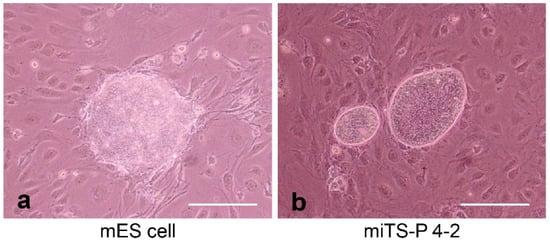
2.1. Induction of Human iTSCs (hiTSCs)
2.2. Induction of hiTSCs from hiPSCs
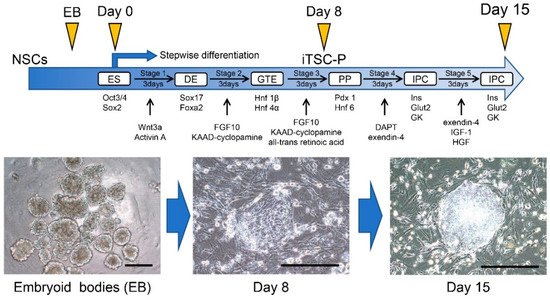
2.3. Induction of IPCs from HDDPC-NSCs
References
- Takahashi, K.; Yamanaka, S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell 2006, 126, 663–676.
- Takahashi, K.; Tanabe, K.; Ohnuki, M.; Narita, M.; Ichisaka, T.; Tomoda, K.; Yamanaka, S. Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell 2007, 131, 861–872.
- Yu, J.; Vodyanik, M.A.; Smuga-Otto, K.; Antosiewicz-Bourget, J.; Frane, J.L.; Tian, S.; Nie, J.; Jonsdottir, G.A.; Ruotti, V.; Stewart, R.; et al. Induced pluripotent stem cell lines derived from human somatic cells. Science 2007, 318, 1917–1920.
- Park, I.H.; Zhao, R.; West, J.A.; Yabuuchi, A.; Huo, H.; Ince, T.A.; Lerou, P.H.; Lensch, M.W.; Daley, G.Q. Reprogramming of human somatic cells to pluripotency with defined factors. Nature 2008, 451, 141–146.
- Maherali, N.; Sridharan, R.; Xie, W.; Utikal, J.; Eminli, S.; Arnold, K.; Stadtfeld, M.; Yachechko, R.; Tchieu, J.; Jaenisch, R.; et al. Directly reprogrammed fibroblasts show global epigenetic remodeling and widespread tissue contribution. Cell Stem Cell 2007, 1, 55–70.
- Okita, K.; Ichisaka, T.; Yamanaka, S. Generation of germline-competent induced pluripotent stem cells. Nature 2007, 448, 313–317.
- Wernig, M.; Meissner, A.; Foreman, R.; Brambrink, T.; Ku, M.; Hochedlinger, K.; Bernstein, B.E.; Jaenisch, R. In vitro reprogramming of fibroblasts into a pluripotent ES-cell-like state. Nature 2007, 448, 318–324.
- Kroon, E.; Martinson, L.A.; Kadoya, K.; Bang, A.G.; Kelly, O.G.; Eliazer, S.; Young, H.; Richardson, M.; Smart, N.G.; Cunningham, J.; et al. Pancreatic endoderm derived from human embryonic stem cells generates glucose-responsive insulin-secreting cells in vivo. Nat. Biotechnol. 2008, 26, 443–452.
- Ohi, Y.; Qin, H.; Hong, C.; Blouin, L.; Polo, J.M.; Guo, T.; Qi, Z.; Downey, S.L.; Manos, P.D.; Rossi, D.J.; et al. Incomplete DNA methylation underlies a transcriptional memory of somatic cells in human iPS cells. Nat. Cell Biol. 2011, 13, 541–549.
- Samavarchi-Tehrani, P.; Golipour, A.; David, L.; Sung, H.K.; Beyer, T.A.; Datti, A.; Woltjen, K.; Nagy, A.; Wrana, J.L. Functional genomics reveals a bmp-driven mesenchymal-to-epithelial transition in the initiation of somatic cell reprogramming. Cell Stem Cell 2010, 7, 64–77.
- Polo, J.M.; Anderssen, E.; Walsh, R.M.; Schwarz, B.A.; Nefzger, C.M.; Lim, S.M.; Borkent, M.; Apostolou, E.; Alaei, S.; Cloutier, J.; et al. A molecular roadmap of reprogramming somatic cells into iPS cells. Cell 2012, 151, 1617–1632.
- Brambrink, T.; Foreman, R.; Welstead, G.G.; Lengner, C.J.; Wernig, M.; Suh, H.; Jaenisch, R. Sequential expression of pluripotency markers during direct reprogramming of mouse somatic cells. Cell Stem Cell 2008, 2, 151–159.
- Stadtfeld, M.; Maherali, N.; Breault, D.T.; Hochedlinger, K. Defining molecular cornerstones during fibroblast to iPS cell reprogramming in mouse. Cell Stem Cell 2008, 2, 230–240.
- Wernig, M.; Lengner, C.J.; Hanna, J.; Lodato, M.A.; Steine, E.; Foreman, R.; Staerk, J.; Markoulaki, S.; Jaenisch, R. A drug-inducible transgenic system for direct reprogramming of multiple somatic cell types. Nat. Biotechnol. 2008, 26, 916–924.
- Stadtfeld, M.; Nagaya, M.; Utikal, J.; Weir, G.; Hochedlinger, K. Induced pluripotent stem cells generated without viral integration. Science 2008, 322, 945–949.
- Fusaki, N.; Ban, H.; Nishiyama, A.; Saeki, K.; Hasegawa, M. Efficient induction of transgene-free human pluripotent stem cells using a vector based on Sendai virus, an RNA virus that does not integrate into the host genome. Proc. Jpn. Acad. Ser. B Phys. Biol. Sci. 2009, 85, 348–862.
- Nishimura, K.; Sano, M.; Ohtaka, M.; Furuta, B.; Umemura, Y.; Nakajima, Y.; Ikehara, Y.; Kobayashi, T.; Segawa, H.; Takayasu, S.; et al. Development of defective and persistent Sendai virus vector: A unique gene delivery/expression system ideal for cell reprogramming. J. Biol. Chem. 2011, 286, 4760–4771.
- Kaji, K.; Norrby, K.; Paca, A.; Mileikovsky, M.; Mohseni, P.; Woltjen, K. Virus-free induction of pluripotency and subsequent excision of reprogramming factors. Nature 2009, 458, 771–775.
- Lahti, A.L.; Kujala, V.J.; Chapman, H.; Koivisto, A.P.; Pekkanen-Mattila, M.; Kerkelä, E.; Hyttinen, J.; Kontula, K.; Swan, H.; Conklin, B.R.; et al. Model for long QT syndrome type 2 using human iPS cells demonstrates arrhythmogenic characteristics in cell culture. Dis. Model. Mech. 2012, 5, 220–230.
- Warren, L.; Manos, P.D.; Ahfeldt, T.; Loh, Y.H.; Li, H.; Lau, F.; Ebina, W.; Mandal, P.K.; Smith, Z.D.; Meissner, A.; et al. Highly efficient reprogramming to pluripotency and directed differentiation of human cells with synthetic modified mRNA. Cell Stem Cell 2010, 7, 618–630.
- Yasuda, S.Y.; Ikeda, T.; Shahsavarani, H.; Yoshida, N.; Nayer, B.; Hino, M.; Vartak-Sharma, N.; Suemori, H.; Hasegawa, K. Chemically defined and growth-factor-free culture system for the expansion and derivation of human pluripotent stem cells. Nat. Biomed. Eng. 2018, 2, 173–182.
- Li, R.; Liang, J.; Ni, S.; Zhou, T.; Qing, X.; Li, H.; He, W.; Chen, J.; Li, F.; Zhuang, Q.; et al. A mesenchymal-to-epithelial transition initiates and is required for the nuclear reprogramming of mouse fibroblasts. Cell Stem Cell 2010, 7, 51–63.
- Panopoulos, A.D.; Yanes, O.; Ruiz, S.; Kida, Y.S.; Diep, D.; Tautenhahn, R.; Herrerías, A.; Batchelder, E.M.; Plongthongkum, N.; Lutz, M.; et al. The metabolome of induced pluripotent stem cells reveals metabolic changes occurring in somatic cell reprogramming. Cell Res. 2012, 22, 168–177.
- Zhang, J.; Nuebel, E.; Daley, G.Q.; Koehler, C.M.; Teitell, M.A. Metabolic regulation in pluripotent stem cells during reprogramming and self-renewal. Cell Stem Cell 2012, 11, 589–595.
- Doege, C.A.; Inoue, K.; Yamashita, T.; Rhee, D.B.; Travis, S.; Fujita, R.; Guarnieri, P.; Bhagat, G.; Vanti, W.B.; Shih, A.; et al. Early-stage epigenetic modification during somatic cell reprogramming by Parp1 and Tet2. Nature 2012, 488, 652–655.
- Krause, M.N.; Sancho-Martinez, I.; Izpisua Belmonte, J.C. Understanding the molecular mechanisms of reprogramming. Biochem. Biophys. Res. Commun. 2016, 473, 693–697.
- Polo, J.M.; Liu, S.; Figueroa, M.E.; Kulalert, W.; Eminli, S.; Tan, K.Y.; Apostolou, E.; Stadtfeld, M.; Li, Y.; Shioda, T.; et al. Cell type of origin influences the molecular and functional properties of mouse induced pluripotent stem cells. Nat. Biotechnol. 2010, 28, 848–855.
- Kim, K.; Doi, A.; Wen, B.; Ng, K.; Zhao, R.; Cahan, P.; Kim, J.; Aryee, M.J.; Ji, H.; Ehrlich, L.I.R.; et al. Epigenetic memory in induced pluripotent stem cells. Nature 2010, 467, 285–290.
- Thiagalingam, S. Epigenetic memory in development and disease: Unraveling the mechanism. Biochim. Biophys. Acta Rev. Cancer 2020, 1873, 188349.
- Thomson, J.A.; Itskovitz-Eldor, J.; Shapiro, S.S.; Waknitz, M.A.; Swiergiel, J.J.; Marshall, V.S.; Jones, J.M. Embryonic stem cell lines derived from human blastocysts. Science 1998, 282, 1145–1147.
- Ilic, D.; Ogilvie, C. Concise review: Human embryonic stem cells-what have we done? What are we doing? Where are we going? Stem Cells 2017, 35, 17–25.
- Tachibana, M.; Amato, P.; Sparman, M.; Gutierrez, N.M.; Tippner-Hedges, R.; Ma, H.; Kang, E.; Fulati, A.; Lee, H.S.; Sritanaudomchai, H.; et al. Human embryonic stem cells derived by somatic cell nuclear transfer. Cell 2013, 153, 1228–1238.
- Chung, Y.G.; Eum, J.H.; Lee, J.E.; Shim, S.H.; Sepilian, V.; Hong, S.W.; Lee, Y.; Treff, N.R.; Choi, Y.H.; Kimbrel, E.A.; et al. Human somatic cell nuclear transfer using adult cells. Cell Stem Cell 2014, 14, 777–780.
- Yamada, M.; Johannesson, B.; Sagi, I.; Burnett, L.C.; Kort, D.H.; Prosser, R.W.; Paull, D.; Nestor, M.W.; Freeby, M.; Greenberg, E.; et al. Human oocytes reprogram adult somatic nuclei of a type 1 diabetic to diploid pluripotent stem cells. Nature 2014, 510, 533–536.
- Wilmut, I.; Schnieke, A.E.; McWhir, J.; Kind, A.J.; Campbell, K.H. Viable offspring derived from fetal and adult mammalian cells. Nature 1997, 385, 810–813.
- Yamanaka, S. Induced pluripotent stem cells: Past, present, and future. Cell Stem Cell 2012, 10, 678–684.
- Fang, F.; Li, Z.; Zhao, Q.; Li, H.; Xiong, C. Human induced pluripotent stem cells and male infertility: An overview of current progress and perspectives. Hum. Reprod. 2018, 33, 188–195.
- Noguchi, H.; Saitoh, I.; Tsugata, T.; Kataoka, H.; Watanabe, M.; Noguchi, Y. Induction of tissue-specific stem cells by reprogramming factors, and tissue-specific selection. Cell Death Differ. 2015, 22, 145–155.
- Bar-Nur, O.; Russ, H.A.; Efrat, S.; Benvenisty, N. Epigenetic memory and preferential lineage-specific differentiation in induced pluripotent stem cells derived from human pancreatic islet beta cells. Cell Stem Cell 2011, 9, 17–23.
- Saitoh, I.; Sato, M.; Soda, M.; Inada, E.; Iwase, Y.; Murakami, T.; Ohshima, H.; Hayasaki, H.; Noguchi, H. Tissue-specific stem cells obtained by reprogramming of non-obese diabetic (nod) mouse-derived pancreatic cells confer insulin production in response to glucose. PLoS ONE 2016, 11, e0163580.
- Mathe, G.; Bernard, J.; Schwarzenberg, L.; Larrieu, M.J.; Lalanne, C.M.; Dutreix, A.; Denoix, P.F.; Surmont, J.; Schwarzmann, V.; Ceoara, B. Trial treatment of patients afflicted with acute leukemia in remission with total irradiation followed by homologous bone marrow transfusion. Rev. Fr. Etud. Clin. Biol. 1959, 4, 675–704.
- Tabatabaei Qomi, R.; Sheykhhasan, M. Adipose-derived stromal cell in regenerative medicine: A review. World J. Stem Cells 2017, 9, 107–117.
- Owczarczyk-Saczonek, A.; Wocior, A.; Placek, W.; Maksymowicz, W.; Wojtkiewicz, J. The use of adipose-derived stem cells in selected skin diseases (vitiligo, alopecia, and nonhealing wounds). Stem Cells Int. 2017, 2017, 4740709.
- Iwazawa, R.; Kozakai, S.; Kitahashi, T.; Nakamura, K.; Hata, K.I. The therapeutic effects of adipose-derived stem cells and recombinant peptide pieces on mouse model of dss colitis. Cell Transplant. 2018, 27, 1390–1400.
- Drozd, A.M.; Walczak, M.P.; Piaskowski, S.; Stoczynska-Fidelus, E.; Rieske, P.; Grzela, D.P. Generation of human iPSCs from cells of fibroblastic and epithelial origin by means of the orip/ebna-1 episomal reprogramming system. Stem Cell Res. Ther. 2015, 6, 122.
- Tamaoki, N.; Takahashi, K.; Tanaka, T.; Ichisaka, T.; Aoki, H.T.K.T.; Takeda-Kawaguchi, T.; Iida, K.; Kunisada, T.; Shibata, T.; Yamanaka, S.; et al. Dental pulp cells for induced pluripotent stem cell banking. J. Dent. Res. 2010, 89, 773–778.
- Yagi, T.; Kosakai, A.; Ito, D.; Okada, Y.; Akamatsu, W.; Nihei, Y.; Nabetani, A.; Ishikawa, F.; Arai, Y.; Hirose, N.; et al. Establishment of induced pluripotent stem cells from centenarians for neurodegenerative disease research. PLoS ONE 2012, 7, e41572.
- Inada, E.; Saitoh, I.; Kubota, N.; Soda, M.; Matsueda, K.; Murakami, T.; Sawami, T.; Kagoshima, A.; Yamasaki, Y.; Sato, M. Alkaline phosphatase and oct-3/4 as useful markers for predicting susceptibility of human deciduous teeth-derived dental pulp cells to reprogramming factor-induced iPS cells. J. Investig. Clin. Dent. 2017, 8.
- Soda, M.; Saitoh, I.; Murakami, T.; Inada, E.; Iwase, Y.; Noguchi, H.; Shibasaki, S.; Kurosawa, M.; Sawami, T.; Terunuma, M.; et al. Repeated human deciduous tooth-derived dental pulp cell reprogramming factor transfection yields multipotent intermediate cells with enhanced iPS cell formation capability. Sci. Rep. 2019, 9, 1490.
- Miyagi-Shiohira, C.; Nakashima, Y.; Kobayashi, N.; Kitamura, S.; Saitoh, I.; Watanabe, M.; Noguchi, H. Induction of expandable adipose-derived mesenchymal stem cells from aged mesenchymal stem cells by a synthetic self-replicating rna. Int. J. Mol. Sci. 2018, 19, 3489.
- Brons, I.G.M.; Smithers, L.E.; Trotter, M.W.; Rugg-Gunn, P.; Sun, B.; de Sousa Lopes, S.M.C.; Howlett, S.K.; Clarkson, A.; Ahrlund-Richter, L.; Pedersen, R.A.; et al. Derivation of pluripotent epiblast stem cells from mammalian embryos. Nature 2007, 448, 191–195.
- Tesar, P.J.; Chenoweth, J.G.; Brook, F.A.; Davies, T.J.; Evans, E.P.; Mack, D.L.; Gardner, R.L.; McKay, R.D. New cell lines from mouse epiblast share defining features with human embryonic stem cells. Nature 2007, 448, 196–199.
- Nichols, J.; Smith, A. Naive and primed pluripotent states. Cell Stem Cell 2009, 4, 487–492.
- Huang, Y.; Osorno, R.; Tsakiridis, A.; Wilson, V. In vivo differentiation potential of epiblast stem cells revealed by chimeric embryo formation. Cell Rep. 2012, 2, 1571–1578.
- Nichols, J.; Smith, A. The origin and identity of embryonic stem cells. Development 2011, 138, 3–8.
- Choi, J.; Lee, S.; Mallard, W.; Clement, K.; Tagliazucchi, G.M.; Lim, H.; Choi, I.Y.; Ferrari, F.; Tsankov, A.M.; Pop, R.; et al. A comparison of genetically matched cell lines reveals the equivalence of human iPSCs and ESCs. Nat. Biotechnol. 2015, 33, 1173–1181.
- Ying, Q.L.; Wray, J.; Nichols, J.; Batlle-Morera, L.; Doble, B.; Woodgett, J.; Cohen, P.; Smith, A. The ground state of embryonic stem cell self-renewal. Nature 2008, 453, 519–523.
- Hanna, J.; Markoulaki, S.; Mitalipova, M.; Cheng, A.W.; Cassady, J.P.; Staerk, J.; Carey, B.W.; Lengner, C.J.; Foreman, R.; Love, J.; et al. Metastable pluripotent states in nod-mouse-derived ESCs. Cell Stem Cell 2009, 4, 513–524.
- Takashima, Y.; Guo, G.; Loos, R.; Nichols, J.; Ficz, G.; Krueger, F.; Oxley, D.; Santos, F.; Clarke, J.; Mansfield, W.; et al. Resetting transcription factor control circuitry toward ground-state pluripotency in human. Cell 2014, 158, 1254–1269.
- Guo, G.; von Meyenn, F.; Santos, F.; Chen, Y.; Reik, W.; Bertone, P.; Smith, A.; Nichols, J. Naive pluripotent stem cells derived directly from isolated cells of the human inner cell mass. Stem Cell Rep. 2016, 6, 437–446.
- Guo, G.; von Meyenn, F.; Rostovskaya, M.; Clarke, J.; Dietmann, S.; Baker, D.; Sahakyan, A.; Myers, S.; Bertone, P.; Reik, W.; et al. Epigenetic resetting of human pluripotency. Development 2017, 144, 2748–2763.
- Hanna, J.; Cheng, A.W.; Saha, K.; Kim, J.; Lengner, C.J.; Soldner, F.; Cassady, J.P.; Muffat, J.; Carey, B.W.; Jaenisch, R. Human embryonic stem cells with biological and epigenetic characteristics similar to those of mouse ESCs. Proc. Natl. Acad. Sci. USA 2010, 107, 9222–9227.
- Inada, E.; Saitoh, I.; Kubota, N.; Iwase, Y.; Murakami, T.; Sawami, T.; Yamasaki, Y.; Sato, M. Increased expression of cell surface ssea-1 is closely associated with naive-like conversion from human deciduous teeth dental pulp cells-derived iPS cells. Int. J. Mol. Sci. 2019, 20, 1651.
- Kiyokawa, Y.; Sato, M.; Noguchi, H.; Inada, E.; Iwase, Y.; Kubota, N.; Sawami, T.; Terunuma, M.; Maeda, T.; Hayasaki, H.; et al. Drug-induced naive iPS cells exhibit better performance than primed iPS cells with respect to the ability to differentiate into pancreatic beta-cell lineage. J. Clin. Med. 2020, 9, 2838.
