SARS-CoV-2 (severe acute respiratory syndrome coronavirus 2) is a virus that belongs to the coronavirus family that provokes respiratory illness in humans. It was transmitted from animals to humans in a mutated form and was first identified in December 2019 in Wuhan, China. The SARS-CoV-2 virus causes COVID-19, a disease named by the World Health Organization on 11 February 2020.
- SARS-CoV-2
- terahertz
- plasmonics
- biosensing
- metasenors
- immunobiosensing
[1][2][3][4][5][6][7][8][9][10][11][12][13][14][15][16][17][18][19][20][21][22][23][24][25][26][27][28][29][30][31][32][33][34][35][36][37][38][39][40][41][42][43][44][45][46][47][48][49][50][51][52][53][54][55][56][57][58][59][60][61][62][63][64][65][66][67][68][69][70][71][72][73][74][75][76][77][78][79][80][81][82][83][84][85][86][87][88][89][90][91][92]
1. Introduction
A rare incidence of pneumonia was identified in China in late 2019 [1], which was later determined to be caused by a novel severe acute respiratory syndrome β-coronavirus (SARS-CoV-2) or novel coronavirus [2]. The World Health Organization (WHO) classified this unique coronavirus disease (COVID-19) as a pandemic after it spread rapidly around the world [3]. There have been 220,563,227 confirmed cases of COVID-19 recorded to WHO as of 6 September 2021, with 4,565,483 deaths and a total of 5,352,927,296 vaccine doses delivered [4]. After severe acute respiratory syndrome (SARS) in 2003 [5] and Middle East Respiratory Syndrome (MERS) in 2012 [6][7][8][6,7,8], COVID-19 is the third large-scale pandemic produced by a coronavirus in the previous two decades. In the case of SARS-CoV-2, the number of laboratory-confirmed COVID-19 infections has now surpassed the total number of SARS and MERS cases by more than 90 times [9]. Despite the new Coronavirus, there are a number of other virus-related diseases that represent a considerable threat to the global population. Therefore, reliable, fast, easy-to-use, and inexpensive virus detection methods are needed to restrain the spread of diseases and preclude future pandemics like COVID-19.
The primary established viral diagnosis methods include CRISPR– Cas12-based detection [10], quantitative real-time polymerase chain reaction (RT-PCR) [11][12][11,12], enzyme-linked immunosorbent assay (ELISA) [13], and point-of-care (POC) lateral flow immunoassay test [14][15][14,15]. Although molecular techniques have been the standard method to detect the presence of viral genetic material in infected patients, it may produce false-negative results if viral RNA is insufficient at the time of detection [16]. Immunoassays [16][17][16,17], which use the spike (S), receptor-binding domain (RBD), and nucleocapsid (N) antigens and IgM, IgG, and IgA antibodies, can provide information on both ongoing viral infections and earlier exposures. The prime drawback of immunoassays is the failure to identify infection during the early stages of the disease, since antibodies take several days to form after contact with foreign material. In [18], a methodological notion for a cell-based biosensor technique has been proposed using membrane-engineered mammalian cells carrying the human chimeric spike S1 antibody. It can produce results within three minutes, with a detection limit of 1 fg/mL and a semi-linear response range of 10 fg to 1μg/mL, albeit clinical validation of the assay using patient samples has yet to be established. Most of these diagnosis processes suffer from some common limitations [16][19][16,19], which include: (i) long turnaround time; (ii) slow detection process; (iii) poor sensitivity; (iv) requirement of certified facilities; (v) need for sophisticated equipment; (vi) well-trained personal to handle the testing; (vii) invasive protocols; and (viii) expense. A detailed summary showing several advantages and disadvantages of these detection methods has been reported in [20].
Several works have recently been reported on plasmonic- and metamaterial-based plasmonic biosensors for virus or viral particle detection, as well as various methods of Coronavirus detection [16][21][22][23][24][16,21,22,23,24], and recent progress in nanophotonic biosensors to combat the COVID-19 pandemic [25]. Although they allow rapid detection with high sensitivity, plasmonic- and metamaterial-based biosensors have not yet become available for clinical use. Considering these available detection methods, Terahertz technology is ideal for addressing these existing detection problems and meeting the constant need for selective, repeatable, label-free, cost-effective, on-chip, ultrasensitive, and easy-to-use biosensors. Terahertz (THz) waves have low energy (~few meV) that is below the ionization energies of atoms and molecules, making it possible to analyze materials without disrupting the system under investigation. The hydrogen bond, the most prominent bond in biological molecules, has characteristic energy within the THz frequency range. Therefore, THz spectroscopy techniques may directly identify spectral properties such as resonances and rotational and vibrational motion of molecules and can be used to detect viruses.
Despite its immense potential, THz-based virus detection is still in its infancy. This paper reviews the existing THz-based virus sensing methods along with COVID-19 sensing and its promising outlook for future pathogenic virus detection to prevent any potential pandemic. The following is the outline of this paper: Section 2 briefly reviews SARS-CoV-2, its existing variants, and available detection methods. In Section 3 , recent advances in virus sensing using terahertz spectroscopy and metamaterials are reviewed. Finally, SARS-CoV-2 detection using terahertz techniques in comparison with conventional methods is presented. The goal of this comprehensive review of terahertz technology for virus detection, including SARS-CoV-2, is to highlight the recent achievements and point out future research opportunities in the field.
2. SARS-CoV-2, Its Variants and Existing Diagnosis Methods
SARS-CoV-2 (severe acute respiratory syndrome coronavirus 2) is a virus that belongs to the coronavirus family that provokes respiratory illness in humans. It was transmitted from animals to humans in a mutated form and was first identified in December 2019 in Wuhan, China. The SARS-CoV-2 virus causes COVID-19, a disease named by the World Health Organization on 11 February 2020 [26]. Coronaviruses (CoVs) are enclosed positive-stranded RNA viruses and possess the largest RNA genome among all known viruses. They belong to the Coronaviridae family which has four primary genera (alpha, beta, gamma, and delta-CoVs) [27][28][27,28].
All CoVs exhibit a similar structure with an identical genome order as shown in Figure 1 [29]. Inside the hemagglutinin esterase viral membrane, the viral genome and nucleocapsid protein (N) bind together to create a helical case. A nucleocapsid (N), a membrane protein (M), a spike protein (S), a small membrane envelope protein (E), and an internal protein (I) are all encoded by the viral gene [28]. One of the initial steps in coronavirus infection is the precise attachment of the coronavirus spike (S) protein to the cellular entry receptors. Several coronavirus receptors have been found [29], including human aminopeptidase N (APN; HCoV-229E), angiotensin-converting enzyme 2 (ACE2; HCoV-NL63, SARS-CoV and SARS-CoV-2), and dipeptidyl peptidase 4 (DPP4; HCoV-NL63, SARS-CoV and SARS-CoV-2) (DPP4; MERS-CoV). As a result, the appearance and the tissue orientation of entrance receptors affect the viral pathogenicity and tropism [30]. It is evident from scanning electron micrographs (SEM) that the coronavirus appears oval or spherical, similar to other coronaviridae family, with stalk-like projections ending in a round structure (spike). The viral particle size ranges from 60 to 140 nm [31][32][31,32] and its infectivity and host specificity are both dependent on spikes.
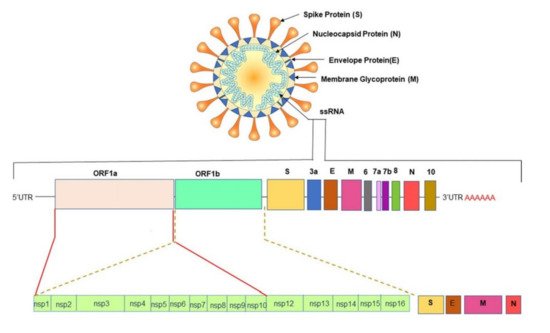
The untranslated regions (5′ and 3′) of large RNA genomes of Coronaviruses contain cis-acting secondary RNA structures which are required for RNA synthesis. Due to the nature of mRNA viruses to make complete duplicates or mutants that are merged into newly made viral particles ( Figure 1 ), Coronaviruses reproduce and express their genomic RNA during their intracellular life cycle. Geographical separation also plays an essential role in producing genetically different variations. The importance of early detection of SARS-CoV-2 has been highlighted by its fast evolution and mutation. New information concerning these variations’ virologic, epidemiologic, and clinical properties is rapidly becoming available. SARS-CoV-2 variants have been identified as 20I/501Y.V1, VOC 202012/01, or B.1.1.7 in the United Kingdom (UK), 20H/501Y.V2 or B.1.351 in South Africa, P.1 in Brazil [33], and E484Q, L245R are two different mutations of B.1.617, which is a double mutant variant. There is also a triple mutant variant B1.618 in India [34][35][36][34,35,36]. Both double and triple mutant variants are now classified as variants of interest (VOIs). There are four distinct mutations in the spike protein of B.1.618 that are linked to enhanced infectivity and immunological escape. New genetic sets have been discovered in the new triple mutant corona virus and parts of the E484 K version are also present in it. The triple mutant corona virus B.1.618 can compromise anyone’s immune system. Antibodies present in the bodies of people who have already been infected with the coronavirus are known as triple mutant corona virus deficient [34]. These new variants are creating considerable concerns due to their capacity to evade natural- or vaccine-induced immunity as well as existing therapies [37][38][37,38]. The new variants are also highly transmissible (70% more transmissible than the old variant), resistant, and carry an elevated risk of death. Efficient testing and detection methods are crucial to suppress the growing infection rate.
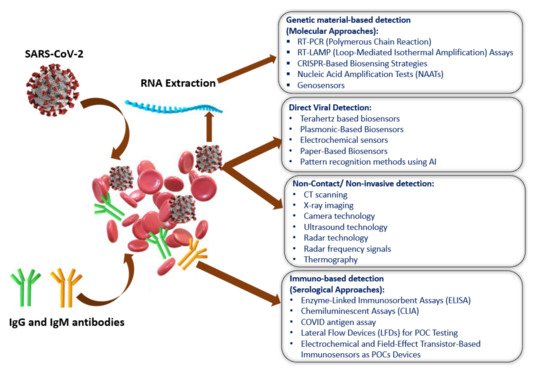
Figure 2 shows the current methods of SARS-CoV-2 detection, which have been categorized into four sections: (i) genetic material-based detection; (ii) direct viral detection; and (iii) non-invasive detection; and (iv) immuno-based detection [39]. The SARS-CoV-2 spike protein is the best option (among the four-protein spike, envelope, matrix, and nucleocapsid) to be used as a diagnostic antigen since it is highly immunogenic and is a key transmembrane protein of the virus. Moreover, the spike protein facilitates the SARS-CoV-2 detection process by revealing the sequence variation of amino acid among coronaviruses.
3. THz Techniques for Virus Sensing
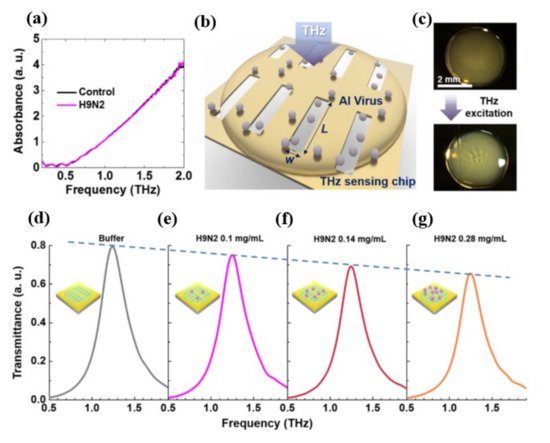
As seen at the top of Figure 35 c, the deposited sample of the virus forms a thin coating on the multi-resonance detecting chip. These nano-antennas were formed by gold with a thickness of 150 nm deposited on a 500-μm-thick double-side-polished silicon wafer. A whole area of 2 mm × 2 mm was designed with more than 1000 slots to reduce the probable errors which could occur during the liquid drop casting because of the random distribution of protein sample. Moreover, each viral sample (H1N1, H5N2) exhibits a distinct transmission spectrum shift as well as a distinct shift in resonance frequency as a result of their different THz absorption characteristics, which indicates the high selectivity potential. According to the authors, the proposed device may identify optically hidden bio specimens, such as viruses, without their distinctive fingerprinting within a dependable spectral span. The optical parameters of the obtained THz spectra for distinct viral samples, including absorption characteristics and complex refractive index, were investigated thoroughly and the tested viruses could be classified based on their subtypes. Furthermore, the viral quantification was successful in a concentration-dependent manner. The transmission spectra of the H9N2 virus were studied using a nano-antenna sensing chip to confirm that the concentration dependent mechanism might allow the THz sensing chip to identify the virus at different concentrations as shown in Figure 35 d–g. Diluting viral samples with buffer liquid in a 1:1 and 2:1 volume ratio yielded four samples with different virus concentrations of 0, 1, 0.14, 0.28 mg/mL, respectively. The maximum transmittance value decreases as the virus concentration rises due to a change in absorption.
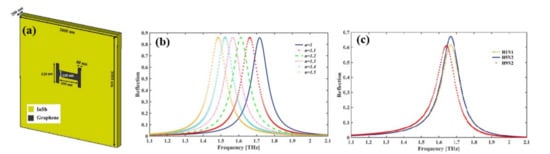
Depending on Fano-resonances a graphene-based H-shaped ( Figure 49 a) nanoscale metamaterial reflector [41][65] positioned at the middle of the InSb semiconductor film was reported for the detection of Avian Influenza (AI) viruses H1N1, H5N2, and H9N2. A resonant frequency shift following the virus deposition, a considerable value of RI sensing (RI FOM up to 2.86) and sensitivity up to 540 GHz/RIU (equivalent to 56.7 μm/RIU) confirms the applicability of the nano bio sensor in virus detection. The reflection spectra of the nanosensor are shown in Figure 49 b with varying RI of the surrounding medium and the spectra blue shifted with decreased RI. The resonant frequencies for H1N1, H5N2, and H9N2 viruses are at 1.668, 1.665, and 1.641 THz, respectively, and the magnitude of the reflection is 61.4%, 67%, and 60.9% respectively ( Figure 49 c).
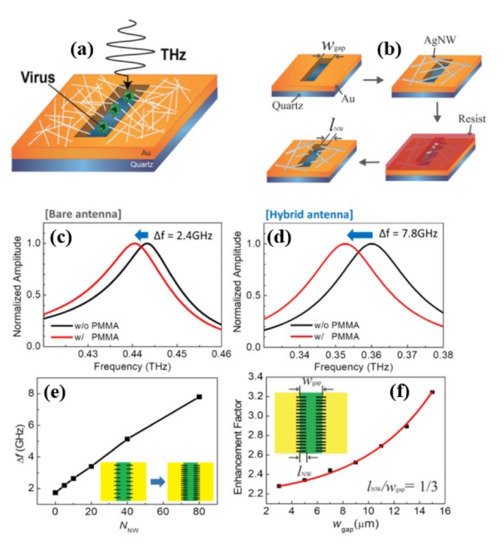
Although nanogap metamaterials [42] was reported recently to improve virus detection limit below 100 nm using the nanolithography process, this fabrication process is difficult, costly, and time-consuming. As a result, one-dimensional nanowires and other novel functional materials with an inherent nanoscale dimension are preferred. In [43][67], silver nanowires (AgNWs) were used to improve the sensitivity of hybrid slot antenna designs for microbial detection in the THz frequency range without using the nanolithography approach as shown in Figure 511 a,b. When poly (methyl methacrylate) (PMMA) and viruses were deposited, a resonant-frequency shift was observed. For a bare slot antenna ( Figure 511 c), resonant frequency exhibits a redshift of 2.4 GHz, whereas a hybrid antenna ( Figure 511 d) indicates a higher frequency shift of 7.8 GHz. Figure 511 f shows Δf as a function of NNW, and it increases with the number density of AgNWs. Figure 511 f shows that the sensitivity enhancement factor is a function of the width of the bare slot antenna. The sensitivity was studied by changing the slot antenna width and the density of the AgNW, and it was found to increase with AgNW density until saturation. An improvement of the sensitivity by a factor of more than 3.8 (~6.7 in terms of FOM) was reported for the 15 μm wide slot antenna. The devices were also used for PRD1 viruses resulting a 2.5 enhancement factor (3.4 in terms of FOM) for the 3 μm slot antenna width. Without using nanoscale fabrication methods, the study demonstrated a nanoscale detecting volume with substantial field enhancement.
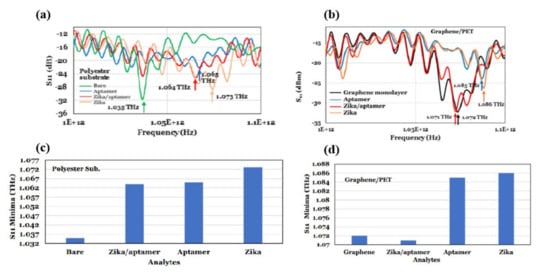
Terahertz reflection spectroscopy without metamaterial substrates was also used to detect Zika virus with and without a targeting/binding oligonucleotides (aptamers) [44][69]. For this study a heat inactivated Zika virus was used. The diameter and mass of the virus was 40 nm and 43 kDa (or 7.1 × 10–20 gm) respectively. In this study, the dielectric substrates were coated with the aptamer and the Zika virus contained pH-neutral buffer solutions in three different combinations for the reflection mode measurements: (i) a 70-µm spacer was utilized to keep the thickness of the Zika/aptamer consistent; (ii) its band gap energy was modified by employing a graphene layer to implement a reflection reference and increase the influence of analyte residual charge; and (iii) a polycarbonate substrate with 0.5 µm microbeads on Zika was used to investigate terahertz properties for various Zika concentrations. The Zika virus had a reflection minimum of 1.073 THz when using terahertz electromagnetic waves (0.75–1.1 THz), whereas Zika/aptamer complexes had a consistent terahertz reflection coefficient minimum of 1.064 THz. The minimum signal detected was of ~16 × 10 3 Zika and the achieved sensitivity was 63 Hz/Zika by measuring the terahertz reflection from polyester microbeads coated with aptamers as a function of Zika concentration, as shown in Figure 613 a,b. The Zika/aptamer combination seems to be performing as a dielectric while utilizing a monolayer of graphene on polyethylene terephthalate (PET), shifting the reflectance minimum from 1.072 GHz for graphene to lower frequency of 1.071 GHz for Zika/aptamer on graphene. As demonstrated in Figure 613 c,d, Zika moved the minimum to 1.085 GHz, while aptamer moved it to 1.086 GHz. Different substrates, including 50 nm-gold films on polycarbonate, 30 nm thin glass slides, and Teflon were also investigated. The authors claim that their sensor could easily detect other viruses, such SARS-CoV-2, by employing specific aptamers or antibodies.
4. THz-Based Sensing and Detections for the Novel SARS-CoV-2
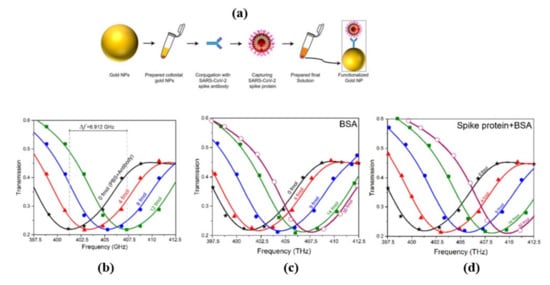
Terahertz toroidal plasmonic metasurfaces can facilitate precise screening and detection of a variety of biomarkers through considerable field confinement at subwavelength geometries [45][46][59,63]. This property paves the way for the detection of targeted biomolecules at low concentrations in extremely diluted solutions. Ahmadivand et al. [47][70] used a toroidal plasmonic metasensor to sense COVID-19 spike protein at femtomolar levels. The reported sensor had a detection limit of ~4.2 fmol and a molecular weight of ~76 kDa, with a process time of around 80 min. The toroidal metasensor performed well at low concentrations, and the shift of the toroidal dipole resonance decreased dramatically and saturated with higher biomolecule concentrations ( Figure 714 ). The feasibility of employing terahertz spectroscopy for early detection of COVID-19 was also investigated in [48][71]. It was demonstrated that viruses with diameters smaller than SARS-COV-2 virus (120 nm) such as PRD1 (60 nm), MS2 (30 nm), Avian Influenza Virus (80–120 nm), Zika Virus (50 nm) can be detected through terahertz technology-based biosensors possessing encouraging sensitivity. These approaches could also be effective to detect coronavirus, and more research into their utility in coronavirus detection is needed. Notable performance comparison of recent biosensors for virus detection are summarized in Table 1 .
| Concept of Work | Sensitivity/LOD | Detected Virus | References |
|---|---|---|---|
| THz toroidal metasensor | ~4.2 fmol | SARS-CoV-2 spike protein | [47] |
| Dual-functional LSPR biosensor with Au Nanoislands | 0.22 pM | SARS-CoV-2 spike protein | [49] |
| Gated graphene-enhanced FET based biosensor | 1.6 × 101 pfu/mL (culture medium) 2.42 × 102 copies/mL (clinical samples) |
SARS-CoV-2 spike protein | [50] |
| Au nanorod based plasmonics | 111.11 deg/RIU | SARS-CoV-2 spike protein | [51] |
| Zr NPs with Zr QDs | 79.15 EID/50 mL | Coronavirus | [52] |
| MIR Spectroscopic technique | 95% | SARS-CoV-2 | [53] |
| Raman Spectroscopic technique | 83.7–97.5% | SARS-CoV-2 | [54] |
| Thermoplasmonic-assisted dual-mode transducing (TP-DMT) | 250 copies/mL | SARS-CoV-2 | [55] |
| LSPR based opto-microfluidic sensing platform with gold nanospikes | 0.08 ng/mL (~0.5 pM) | SARS-CoV-2 | [56] |
| Carboxyl groups (PC)-coated magnetic nanoparticles (pcMNPs) | 10 copy/mL | SARS-CoV-2 RNA | [57] |
| THz Slot antenna with silver nanowire | 32.7 GHz·µm2/particle | PRD1 | [43] |
| THz Toroidal metasensor | 5.81 GHz/log(pg/mL) | Zika virus | [46] |
| THz Toroidal metasensor | 6.47 GHz/log (pg/mL) | Zika virus | [45] |
| THz Au nano-antenna with 2D punctured rectangular slots | 0.35 THz/RIU | H1N1, H9N2, H5N2 | [40] |
| THz Rectangular Au metamaterial | 80 GHz/particle/µm2 | PRD1, MS2 | [42] |
| THz SSPP Jerusalem Cross Aperture | 0.5 THz/RIU | AIV | [58] |
| Hetero-assembled AuNPs sandwich-immunoassay LSPR chip | 100 fg mL−1 | Hepatitis B surface antigen | [59] |
References
- Huang, C.; Wang, Y.; Li, X.; Ren, L.; Zhao, J.; Hu, Y.; Zhang, L.; Fan, G.; Xu, J.; Gu, X.; et al. Clinical Features of Patients Infected with 2019 Novel Coronavirus in Wuhan, China. Lancet 2020, 395, 497–506. Huang, C.; Wang, Y.; Li, X.; Ren, L.; Zhao, J.; Hu, Y.; Zhang, L.; Fan, G.; Xu, J.; Gu, X.; et al. Clinical Features of Patients Infected with 2019 Novel Coronavirus in Wuhan, China. Lancet 2020, 395, 497–506, doi:10.1016/S0140-6736(20)30183-5.
- Lu, R.; Zhao, X.; Li, J.; Niu, P.; Yang, B.; Wu, H.; Wang, W.; Song, H.; Huang, B.; Zhu, N.; et al. Genomic Characterisation and Epidemiology of 2019 Novel Coronavirus: Implications for Virus Origins and Receptor Binding. Lancet 2020, 395, 565–574. Lu, R.; Zhao, X.; Li, J.; Niu, P.; Yang, B.; Wu, H.; Wang, W.; Song, H.; Huang, B.; Zhu, N.; et al. Genomic Characterisation and Epidemiology of 2019 Novel Coronavirus: Implications for Virus Origins and Receptor Binding. Lancet 2020, 395, 565–574, doi:10.1016/S0140-6736(20)30251-8.
- Cucinotta, D.; Vanelli, M. WHO Declares COVID-19 a Pandemic. Acta Biomed. 2020, 91, 157–160. Cucinotta, D.; Vanelli, M. WHO Declares COVID-19 a Pandemic. Acta Biomed. 2020, 91, 157–160.
- WHO Coronavirus (COVID-19) Dashboard | WHO Coronavirus (COVID-19) Dashboard with Vaccination Data. Available online: https://covid19.who.int/ (accessed on 7 September 2021).
- Zhong, N.S.; Zheng, B.J.; Li, Y.M.; Poon, L.L.M.; Xie, Z.H.; Chan, K.H.; Li, P.H.; Tan, S.Y.; Chang, Q.; Xie, J.P.; et al. Epidemiology and Cause of Severe Acute Respiratory Syndrome (SARS) in Guangdong, People’s Republic of China, in February, 2003. Lancet 2003, 362, 1353–1358. Zhong, N.S.; Zheng, B.J.; Li, Y.M.; Poon, L.L.M.; Xie, Z.H.; Chan, K.H.; Li, P.H.; Tan, S.Y.; Chang, Q.; Xie, J.P.; et al. Epidemiology and Cause of Severe Acute Respiratory Syndrome (SARS) in Guangdong, People’s Republic of China, in February, 2003. Lancet 2003, 362, 1353–1358, doi:10.1016/S0140-6736(03)14630-2.
- Alhamlan, F.S.; Majumder, M.S.; Brownstein, J.S.; Hawkins, J.; Al-Abdely, H.M.; Alzahrani, A.; Obaid, D.A.; Al-Ahdal, M.N.; BinSaeed, A. Case Characteristics among Middle East Respiratory Syndrome Coronavirus Outbreak and Non-Outbreak Cases in Saudi Arabia from 2012 to 2015. BMJ Open 2017, 7, 11865. Alhamlan, F.S.; Majumder, M.S.; Brownstein, J.S.; Hawkins, J.; Al-Abdely, H.M.; Alzahrani, A.; Obaid, D.A.; Al-Ahdal, M.N.; BinSaeed, A. Case Characteristics among Middle East Respiratory Syndrome Coronavirus Outbreak and Non-Outbreak Cases in Saudi Arabia from 2012 to 2015. BMJ Open 2017, 7, 11865, doi:10.1136/bmjopen-2016-011865.
- Wang, L.F.; Anderson, D.E. Viruses in Bats and Potential Spillover to Animals and Humans. Curr. Opin. Virol. 2019, 34, 79–89. Wang, L.F.; Anderson, D.E. Viruses in Bats and Potential Spillover to Animals and Humans. Curr. Opin. Virol. 2019, 34, 79–89.
- Ksiazek, T.G.; Erdman, D.; Goldsmith, C.S.; Zaki, S.R.; Peret, T.; Emery, S.; Tong, S.; Urbani, C.; Comer, J.A.; Lim, W.; et al. A Novel Coronavirus Associated with Severe Acute Respiratory Syndrome. N. Engl. J. Med. 2003, 348, 1953–1966. Ksiazek, T.G.; Erdman, D.; Goldsmith, C.S.; Zaki, S.R.; Peret, T.; Emery, S.; Tong, S.; Urbani, C.; Comer, J.A.; Lim, W.; et al. A Novel Coronavirus Associated with Severe Acute Respiratory Syndrome. N. Engl. J. Med. 2003, 348, 1953–1966, doi:10.1056/nejmoa030781.
- Peeri, N.C.; Shrestha, N.; Siddikur Rahman, M.; Zaki, R.; Tan, Z.; Bibi, S.; Baghbanzadeh, M.; Aghamohammadi, N.; Zhang, W.; Haque, U. The SARS, MERS and Novel Coronavirus (COVID-19) Epidemics, the Newest and Biggest Global Health Threats: What Lessons Have We Learned? Int. J. Epidemiol. 2021, 49, 717–726. Peeri, N.C.; Shrestha, N.; Siddikur Rahman, M.; Zaki, R.; Tan, Z.; Bibi, S.; Baghbanzadeh, M.; Aghamohammadi, N.; Zhang, W.; Haque, U. The SARS, MERS and Novel Coronavirus (COVID-19) Epidemics, the Newest and Biggest Global Health Threats: What Lessons Have We Learned? Int. J. Epidemiol. 2021, 49, 717–726.
- Broughton, J.P.; Deng, X.; Yu, G.; Fasching, C.L.; Servellita, V.; Singh, J.; Miao, X.; Streithorst, J.A.; Granados, A.; Sotomayor-Gonzalez, A.; et al. CRISPR–Cas12-Based Detection of SARS-CoV-2. Nat. Biotechnol. 2020, 38, 870–874. Broughton, J.P.; Deng, X.; Yu, G.; Fasching, C.L.; Servellita, V.; Singh, J.; Miao, X.; Streithorst, J.A.; Granados, A.; Sotomayor-Gonzalez, A.; et al. CRISPR–Cas12-Based Detection of SARS-CoV-2. Nat. Biotechnol. 2020, 38, 870–874, doi:10.1038/s41587-020-0513-4.
- Pan, Y.; Zhang, D.; Yang, P.; Poon, L.L.M.; Wang, Q. Viral Load of SARS-CoV-2 in Clinical Samples. Lancet Infect. Dis. 2020, 20, 411–412. Pan, Y.; Zhang, D.; Yang, P.; Poon, L.L.M.; Wang, Q. Viral Load of SARS-CoV-2 in Clinical Samples. Lancet Infect. Dis. 2020, 20, 411–412.
- Li, Y.; Yao, L.; Li, J.; Chen, L.; Song, Y.; Cai, Z.; Yang, C. Stability Issues of RT-PCR Testing of SARS-CoV-2 for Hospitalized Patients Clinically Diagnosed with COVID-19. J. Med. Virol. 2020, 92, 903–908. Li, Y.; Yao, L.; Li, J.; Chen, L.; Song, Y.; Cai, Z.; Yang, C. Stability Issues of RT-PCR Testing of SARS-CoV-2 for Hospitalized Patients Clinically Diagnosed with COVID-19. J. Med. Virol. 2020, 92, 903–908, doi:10.1002/jmv.25786.
- Adams, E.; Ainsworth, M.; Anand, R.; Andersson, M.I.; Auckland, K.; Baillie, J.K.; Barnes, E.; Beer, S.; Bell, J.I.; Berry, T.; et al. Antibody Testing for COVID-19: A Report from the National COVID Scientific Advisory Panel. Wellcome Open Res. 2020, 5, 139. Adams, E.; Ainsworth, M.; Anand, R.; Andersson, M.I.; Auckland, K.; Baillie, J.K.; Barnes, E.; Beer, S.; Bell, J.I.; Berry, T.; et al. Antibody Testing for COVID-19: A Report from the National COVID Scientific Advisory Panel. Wellcome Open Research 2020, 5, 139, doi: 10.12688/wellcomeopenres.15927.1.
- Nguyen, T.; Bang, D.D.; Wolff, A. 2019 Novel Coronavirus Disease (COVID-19): Paving the Road for Rapid Detection and Point-of-Care Diagnostics. Micromachines 2020, 11, 306. Nguyen, T.; Bang, D.D.; Wolff, A. 2019 Novel Coronavirus Disease (COVID-19): Paving the Road for Rapid Detection and Point-of-Care Diagnostics. Micromachines 2020, 11, 1–7, doi:10.3390/MI11030306.
- Visseaux, B.; le Hingrat, Q.; Collin, G.; Bouzid, D.; Lebourgeois, S.; le Pluart, D.; Deconinck, L.; Lescure, F.X.; Lucet, J.C.; Bouadma, L.; et al. Evaluation of the Qiastat-Dx Respiratory Sars-Cov-2 Panel, the First Rapid Multiplex PCR Commercial Assay for Sars-Cov-2 Detection. J. Clin. Microbiol. 2020, 58, e00630-20. Visseaux, B.; le Hingrat, Q.; Collin, G.; Bouzid, D.; Lebourgeois, S.; le Pluart, D.; Deconinck, L.; Lescure, F.X.; Lucet, J.C.; Bouadma, L.; et al. Evaluation of the Qiastat-Dx Respiratory Sars-Cov-2 Panel, the First Rapid Multiplex PCR Commercial Assay for Sars-Cov-2 Detection. J. Clin. Microbiol. 2020, 58, e00630-20 , doi:10.1128/JCM.00630-20.
- Ji, T.; Liu, Z.; Wang, G.Q.; Guo, X.; Akbar khan, S.; Lai, C.; Chen, H.; Huang, S.; Xia, S.; Chen, B.; et al. Detection of COVID-19: A Review of the Current Literature and Future Perspectives. Biosens. Bioelectron. 2020, 166, 112455. Ji, T.; Liu, Z.; Wang, G.Q.; Guo, X.; Akbar khan, S.; Lai, C.; Chen, H.; Huang, S.; Xia, S.; Chen, B.; et al. Detection of COVID-19: A Review of the Current Literature and Future Perspectives. Biosens. Bioelectron. 2020, 166, 112455, doi:10.1016/J.BIOS.2020.112455.
- Lee, C.Y.-P.; Lin, R.T.P.; Renia, L.; Ng, L.F.P. Serological Approaches for COVID-19: Epidemiologic Perspective on Surveillance and Control. Front. Immunol. 2020, 11, 879. Lee, C.Y.-P.; Lin, R.T.P.; Renia, L.; Ng, L.F.P. Serological Approaches for COVID-19: Epidemiologic Perspective on Surveillance and Control. Front. Immunol. 2020, 11, 879, doi:10.3389/FIMMU.2020.00879.
- Mavrikou, S.; Moschopoulou, G.; Tsekouras, V.; Kintzios, S. Development of a Portable, Ultra-Rapid and Ultra-Sensitive Cell-Based Biosensor for the Direct Detection of the SARS-CoV-2 S1 Spike Protein Antigen. Sensors 2020, 20, 3121. Mavrikou, S.; Moschopoulou, G.; Tsekouras, V.; Kintzios, S. Development of a Portable, Ultra-Rapid and Ultra-Sensitive Cell-Based Biosensor for the Direct Detection of the SARS-CoV-2 S1 Spike Protein Antigen. Sensors 2020, 20, 3121, doi:10.3390/S20113121.
- Tang, Y.W.; Schmitz, J.E.; Persing, D.H.; Stratton, C.W. Laboratory Diagnosis of COVID-19: Current Issues and Challenges. J. Clin. Microbiol. 2020, 58, e00512-20. Tang, Y.W.; Schmitz, J.E.; Persing, D.H.; Stratton, C.W. Laboratory Diagnosis of COVID-19: Current Issues and Challenges. J. Clin. Microbiol. 2020, 58, e00512-20.
- Eftekhari, A.; Alipour, M.; Chodari, L.; Dizaj, S.M.; Ardalan, M.; Samiei, M.; Sharifi, S.; Vahed, S.Z.; Huseynova, I.; Khalilov, R.; et al. A Comprehensive Review of Detection Methods for SARS-CoV-2. Microorganisms 2021, 9, 232. Eftekhari, A.; Alipour, M.; Chodari, L.; Dizaj, S.M.; Ardalan, M.; Samiei, M.; Sharifi, S.; Vahed, S.Z.; Huseynova, I.; Khalilov, R.; et al. A Comprehensive Review of Detection Methods for SARS-CoV-2. Microorganisms 2021, 9, 232, doi:10.3390/MICROORGANISMS9020232.
- Mauriz, E. Recent Progress in Plasmonic Biosensing Schemes for Virus Detection. Sensors 2020, 20, 4745. Mauriz, E. Recent Progress in Plasmonic Biosensing Schemes for Virus Detection. Sensors 2020, 20, 4745, doi:10.3390/S20174745.
- Samson, R.; Navale, G.R.; Dharne, M.S. Biosensors: Frontiers in Rapid Detection of COVID-19. 3 Biotech 2020, 10, 1–9. Samson, R.; Navale, G.R.; Dharne, M.S. Biosensors: Frontiers in Rapid Detection of COVID-19. 3 Biotech 2020, 10, 1–9, doi:10.1007/S13205-020-02369-0.
- Yüce, M.; Filiztekin, E.; Özkaya, K.G. COVID-19 Diagnosis—A Review of Current Methods. Biosens. Bioelectron. 2021, 172, 112752. Yüce, M.; Filiztekin, E.; Özkaya, K.G. COVID-19 Diagnosis —A Review of Current Methods. Biosens. Bioelectron. 2021, 172, 112752, doi:10.1016/J.BIOS.2020.112752.
- Antiochia, R. Developments in Biosensors for CoV Detection and Future Trends. Biosens. Bioelectron. 2021, 173, 112777. Antiochia, R. Developments in Biosensors for CoV Detection and Future Trends. Biosens. Bioelectron. 2021, 173, 112777, doi:10.1016/J.BIOS.2020.112777.
- Soler, M.; Estevez, M.C.; Cardenosa-Rubio, M.; Astua, A.; Lechuga, L.M. How Nanophotonic Label-Free Biosensors Can Contribute to Rapid and Massive Diagnostics of Respiratory Virus Infections: COVID-19 Case. ACS Sens. 2020, 5, 2663–2678. Soler, M.; Estevez, M.C.; Cardenosa-Rubio, M.; Astua, A.; Lechuga, L.M. How Nanophotonic Label-Free Biosensors Can Contribute to Rapid and Massive Diagnostics of Respiratory Virus Infections: COVID-19 Case. ACS Sens. 2020, 5, 2663–2678, doi:10.1021/ACSSENSORS.0C01180.
- Naming the Coronavirus Disease (COVID-19) and the Virus That Causes It. Available online: https://https://www.who.int/emergencies/diseases/novel-coronavirus-2019/technical-guidance/naming-the-coronavirus-disease-(covid-2019)-and-the-virus-that-causes-it (accessed on 18 June 2021).Naming the Coronavirus Disease (COVID-19) and the Virus That Causes It. Available online: https://www.who.int/emergencies/diseases/novel-coronavirus-2019/technical-guidance/naming-the-coronavirus-disease-(covid-2019)-and-the-virus-that-causes-it (accessed on 18 June 2021).
- Corman, V.M.; Muth, D.; Niemeyer, D.; Drosten, C. Hosts and Sources of Endemic Human Coronaviruses. In Advances in Virus Research; Academic Press Inc.: Cambridge, MA, USA, 2018; Volume 100, pp. 163–188. ISBN 9780128152010. Corman, V.M.; Muth, D.; Niemeyer, D.; Drosten, C. Hosts and Sources of Endemic Human Coronaviruses. In Advances in Virus Research; Academic Press Inc.: Cambridge, MA, USA 2018; Volume 100, pp. 163–188; ISBN 9780128152010.
- Weiss, S.R.; Leibowitz, J.L. Coronavirus pathogenesis. In Advances in Virus Research; Academic Press Inc.: Cambridge, MA, USA, 2011; Volume 81, pp. 85–164. Weiss, S.R.; Leibowitz, J.L. Coronavirus pathogenesis. In Advances in Virus Research; Academic Press Inc.: Cambridge, MA, USA 2011; Volume 81, pp. 85–164.
- Rastogi, M.; Pandey, N.; Shukla, A.; Singh, S.K. SARS Coronavirus 2: From Genome to Infectome. Respir. Res. 2020, 21, 1–15. Rastogi, M.; Pandey, N.; Shukla, A.; Singh, S.K. SARS Coronavirus 2: From Genome to Infectome. Respir. Res. 2020, 21, 1–15.
- V’kovski, P.; Kratzel, A.; Steiner, S.; Stalder, H.; Thiel, V. Coronavirus Biology and Replication: Implications for SARS-CoV-2. Nat. Rev. Microbiol. 2021, 19, 155–170. V’kovski, P.; Kratzel, A.; Steiner, S.; Stalder, H.; Thiel, V. Coronavirus Biology and Replication: Implications for SARS-CoV-2. Nat. Rev. Microbiol. 2021, 19, 155–170.
- Varga, Z.; Flammer, A.J.; Steiger, P.; Haberecker, M.; Andermatt, R.; Zinkernagel, A.; Mehra, M.R.; Scholkmann, F.; SchÃ, R.; Ruschitzka, F.; et al. Electron Microscopy of SARS-CoV-2: A Challenging Task–Authors’ Reply. Lancet 2020, 395, e100. Varga, Z.; Flammer, A.J.; Steiger, P.; Haberecker, M.; Andermatt, R.; Zinkernagel, A.; Mehra, M.R.; Scholkmann, F.; SchÃ, R.; Ruschitzka, F.; et al. Electron Microscopy of SARS-CoV-2: A Challenging Task–Authors’ Reply. Lancet 2020, 395, e100, doi:10.1016/j.kint.2020.04.003.
- Zhu, N.; Zhang, D.; Wang, W.; Li, X.; Yang, B.; Song, J.; Zhao, X.; Huang, B.; Shi, W.; Lu, R.; et al. A Novel Coronavirus from Patients with Pneumonia in China, 2019. N. Engl. J. Med. 2020, 382, 727–733. Zhu, N.; Zhang, D.; Wang, W.; Li, X.; Yang, B.; Song, J.; Zhao, X.; Huang, B.; Shi, W.; Lu, R.; et al. A Novel Coronavirus from Patients with Pneumonia in China, 2019. N. Engl. J. Med. 2020, 382, 727–733, doi:10.1056/nejmoa2001017.
- Science Brief: Emerging SARS-CoV-2 Variants | CDC. Available online: https://www.cdc.gov/coronavirus/2019-ncov/science/science-briefs/scientific-brief-emerging-variants.html (accessed on 19 June 2021).
- Triple Mutant Corona Virus Found in India, More Impact in Maharashtra and Bengal. Available online: https://www.zoomnews.in/en/news-detail/triple-mutant-corona-virus-found-in-india-more-impact-in-maharashtra-and-bengal-1.html (accessed on 19 June 2021).
- Indian Covid-19 Variant (B.1.617) | New Scientist. Available online: https://www.newscientist.com/definition/indian-covid-19-variant-b-1-617/ (accessed on 19 June 2021).
- WHO Classifies Triple-Mutant Covid Variant from India as Global Health Risk. Available online: https://www.cnbc.com/2021/05/10/who-classifies-triple-mutant-covid-variant-from-india-as-global-health-risk-.html (accessed on 19 June 2021).
- Rauseo, A.M.; O’Halloran, J.A. What Are the Clinical Implications of the SARS-CoV-2 Variants: 5 Things Every Cardiologist Should Know. JACC: Basic Transl. Sci. 2021, 6, 305–308. Rauseo, A.M.; O’Halloran, J.A. What Are the Clinical Implications of the SARS-CoV-2 Variants: 5 Things Every Cardiologist Should Know. JACC: Basic Transl. Sci. 2021, 6, 305–308, doi:10.1016/j.jacbts.2021.02.009.
- Wu, K.; Werner, A.P.; Moliva, J.I.; Koch, M.; Choi, A.; Stewart-Jones, G.B.E.; Bennett, H.; Boyoglu-Barnum, S.; Shi, W.; Graham, B.S.; et al. MRNA-1273 Vaccine Induces Neutralizing Antibodies against Spike Mutants from Global SARS-CoV-2 Variants. bioRxiv 2021. Wu, K.; Werner, A.P.; Moliva, J.I.; Koch, M.; Choi, A.; Stewart-Jones, G.B.E.; Bennett, H.; Boyoglu-Barnum, S.; Shi, W.; Graham, B.S.; et al. MRNA-1273 Vaccine Induces Neutralizing Antibodies against Spike Mutants from Global SARS-CoV-2 Variants. bioRxiv 2021, doi:10.1101/2021.01.25.427948.
- Mattioli, I.A.; Hassan, A.; Oliveira, O.N.; Crespilho, F.N. On the Challenges for the Diagnosis of SARS-CoV-2 Based on a Review of Current Methodologies. ACS Sens. 2020, 5, 3655–3677. Mattioli, I.A.; Hassan, A.; Oliveira, O.N.; Crespilho, F.N. On the Challenges for the Diagnosis of SARS-CoV-2 Based on a Review of Current Methodologies. ACS Sens. 2020, 5, 3655–3677, doi:10.1021/acssensors.0c01382.
- Lee, D.-K.; Kang, J.-H.; Kwon, J.; Lee, J.-S.; Lee, S.; Woo, D.H.; Kim, J.H.; Song, C.-S.; Park, Q.-H.; Seo, M. Nano Metamaterials for Ultrasensitive Terahertz Biosensing. Sci. Rep. 2017, 7, 1–6. Yang, X.; Zhao, X.; Yang, K.; Liu, Y.; Liu, Y.; Fu, W.; Luo, Y. Biomedical Applications of Terahertz Spectroscopy and Imaging. Trends Biotechnol. 2016, 34, 810–824.
- Keshavarz, A.; Vafapour, Z. Sensing Avian Influenza Viruses Using Terahertz Metamaterial Reflector. IEEE Sens. J. 2019, 19, 5161–5166. Uddin, J. (Ed.) Terahertz Spectroscopy-A Cutting Edge Technology; IntechOpen,, 2017; doi:10.5772/67031. Available online: https://www.intechopen.com/chapters/54052 (accessed on 19 June 2021).
- Park, S.J.; Cha, S.H.; Shin, G.A.; Ahn, Y.H. Sensing Viruses Using Terahertz Nano-Gap Metamaterials. Biomed. Opt. Express 2017, 8, 3551. Park, S.J.; Cha, S.H.; Shin, G.A.; Ahn, Y.H. Sensing Viruses Using Terahertz Nano-Gap Metamaterials. Biomed. Opt. Express 2017, 8, 3551, doi:10.1364/boe.8.003551.
- Hong, J.T.; Jun, S.W.; Cha, S.H.; Park, J.Y.; Lee, S.; Shin, G.A.; Ahn, Y.H. Enhanced Sensitivity in THz Plasmonic Sensors with Silver Nanowires. Sci. Rep. 2018, 8, 15536. Lee, Y.S. Principles of Terahertz Science and Technology; Springer: New York, NY, USA, 2009; ISBN 9780387095394.
- Dolai, S.; Tabib-Azar, M. Terahertz Detection of Zika Viruses. Preprints 2020, 2020020232. Ajito, K.; Ueno, Y. THz Chemical Imaging for Biological Applications. IEEE Trans. Terahertz Sci. Technol. 2011, 1, 293–300, doi:10.1109/TTHZ.2011.2159562.
- Ahmadivand, A.; Gerislioglu, B.; Manickam, P.; Kaushik, A.; Bhansali, S.; Nair, M.; Pala, N. Rapid Detection of Infectious Envelope Proteins by Magnetoplasmonic Toroidal Metasensors. ACS Sens. 2017, 2, 1359–1368. Son, J.H.; Oh, S.J.; Cheon, H. Potential Clinical Applications of Terahertz Radiation. J. Appl. Phys. 2019, 125, 190901.
- Ahmadivand, A.; Gerislioglu, B.; Tomitaka, A.; Manickam, P.; Kaushik, A.; Bhansali, S.; Nair, M.; Pala, N. Extreme Sensitive Metasensor for Targeted Biomarkers Identification Using Colloidal Nanoparticles-Integrated Plasmonic Unit Cells. Biomed. Opt. Express 2018, 9, 373. Lu, F.; Ou, H.; Liao, Y.; Zhu, F.; Lin, Y.S. Actively Switchable Terahertz Metamaterial. Results Phys. 2019, 15, 102756, doi:10.1016/J.RINP.2019.102756.
- Ahmadivand, A.; Gerislioglu, B.; Ramezani, Z.; Kaushik, A.; Manickam, P.; Ghoreishi, S.A. Functionalized Terahertz Plasmonic Metasensors: Femtomolar-Level Detection of SARS-CoV-2 Spike Proteins. Biosens. Bioelectron. 2021, 177, 112971. Di Fabrizio, M.; Lupi, S.; D’Arco, A. Virus Recognition with Terahertz Radiation: Drawbacks and Potentialities. J. Phys. Photonics 2021, 3, 32001, doi:10.1088/2515-7647/abfd08.
- Terahertz Technology for the Identification of the COVID-19. Review of Academic Literature March 2020, TERAGROUP Prepared in Cooperation with: The Israeli Ministry of Defense & Rabatit Engineering & Consulting Ltd. Available online: https://www.iclinics.cl/wp-content/uploads/2020/10/tae-Terahertz-Technology-for-the-Identification-of-the-COVID-19-Academic-Literature.pdf (accessed on 19 June 2021).Seo, M.; Park, H.-R. Terahertz Biochemical Molecule-Specific Sensors. Adv. Opt. Mater. 2020, 8, 1900662, doi:10.1002/ADOM.201900662.
- Qiu, G.; Gai, Z.; Tao, Y.; Schmitt, J.; Kullak-Ublick, G.A.; Wang, J. Dual-Functional Plasmonic Photothermal Biosensors for Highly Accurate Severe Acute Respiratory Syndrome Coronavirus 2 Detection. ACS Nano 2020, 14, 5268–5277. Lee, D.-K.; Kang, J.-H.; Kwon, J.; Lee, J.-S.; Lee, S.; Woo, D.H.; Kim, J.H.; Song, C.-S.; Park, Q.-H.; Seo, M. Nano Metamaterials for Ultrasensitive Terahertz Biosensing. Sci. Rep. 2017, 7, 1–6, doi:10.1038/s41598-017-08508-7.
- Seo, G.; Lee, G.; Kim, M.J.; Baek, S.-H.; Choi, M.; Ku, K.B.; Lee, C.-S.; Jun, S.; Park, D.; Kim, H.G.; et al. Rapid Detection of COVID-19 Causative Virus (SARS-CoV-2) in Human Nasopharyngeal Swab Specimens Using Field-Effect Transistor-Based Biosensor. ACS Nano 2020, 14, 5135–5142. Acbas, G.; Niessen, K.A.; Snell, E.H.; Markelz, A.G. Optical Measurements of Long-Range Protein Vibrations. Nat. Commun. 2014, 5, 1–7, doi:10.1038/ncomms4076.
- Das, C.M.; Guo, Y.; Yang, G.; Kang, L.; Xu, G.; Ho, H.P.; Yong, K.T. Gold Nanorod Assisted Enhanced Plasmonic Detection Scheme of COVID-19 SARS-CoV-2 Spike Protein. Adv. Theory Simul. 2020, 3, 2000185. Park, H.R.; Park, Y.M.; Kim, H.S.; Kyoung, J.S.; Seo, M.A.; Park, D.J.; Ahn, Y.H.; Ahn, K.J.; Kim, D.S. Terahertz Nanoresonators: Giant Field Enhancement and Ultrabroadband Performance. Appl. Phys. Lett. 2010, 96, 121106, doi:10.1063/1.3368690.
- Ahmed, S.R.; Kang, S.W.; Oh, S.; Lee, J.; Neethirajan, S. Chiral Zirconium Quantum Dots: A New Class of Nanocrystals for Optical Detection of Coronavirus. Heliyon 2018, 4, e00766. Cheng, D.; He, X.; Huang, X.; Zhang, B.; Liu, G.; Shu, G.; Fang, C.; Wang, J.; Luo, Y. Terahertz Biosensing Metamaterial Absorber for Virus Detection Based on Spoof Surface Plasmon Polaritons. Int. J. RF Microw. Comput.-Aided Eng. 2018, 28, e21448, doi:10.1002/mmce.21448
- Barauna, V.G.; Singh, M.N.; Barbosa, L.L.; Marcarini, W.D.; Vassallo, P.F.; Mill, J.G.; Ribeiro-Rodrigues, R.; Campos, L.C.G.; Warnke, P.H.; Martin, F.L. Ultrarapid On-Site Detection of SARS-CoV-2 Infection Using Simple ATR-FTIR Spectroscopy and an Analysis Algorithm: High Sensitivity and Specificity. Anal. Chem. 2021, 93, 2950–2958. Ou, H.; Lu, F.; Xu, Z.; Lin, Y.-S. Terahertz Metamaterial with Multiple Resonances for Biosensing Application. Nanomaterials 2020, 10, 1038, doi:10.3390/NANO10061038.
- Carlomagno, C.; Bertazioli, D.; Gualerzi, A.; Picciolini, S.; Banfi, P.I.; Lax, A.; Messina, E.; Navarro, J.; Bianchi, L.; Caronni, A.; et al. COVID-19 Salivary Raman Fingerprint: Innovative Approach for the Detection of Current and Past SARS-CoV-2 Infections. Sci. Rep. 2021, 11, 1–13. Zhou, H.; Yang, C.; Hu, D.; Li, D.; Hui, X.; Zhang, F.; Chen, M.; Mu, X. Terahertz Biosensing Based on Bi-Layer Metamaterial Absorbers toward Ultra-High Sensitivity and Simple Fabrication. Appl. Phys. Lett. 2019, 115, 143507, doi:10.1063/1.5111584.
- Qiu, G.; Gai, Z.; Saleh, L.; Tang, J.; Gui, T.; Kullak-Ublick, G.A.; Wang, J. Thermoplasmonic-Assisted Cyclic Cleavage Amplification for Self-Validating Plasmonic Detection of SARS-CoV-2. ACS Nano 2021, 15, 7536–7546. Tan, S.; Yan, F.; Wang, W.; Zhou, H.; Hou, Y. Ultrasensitive Sensing with Three-Dimensional Terahertz Metamaterial Absorber. J. Opt. 2018, 20, 055101, doi:10.1088/2040-8986/AAB66E.
- Funari, R.; Chu, K.Y.; Shen, A.Q. Detection of Antibodies against SARS-CoV-2 Spike Protein by Gold Nanospikes in an Opto-Microfluidic Chip. Biosens. Bioelectron. 2020, 169, 112578. Hu, X.; Xu, G.; Wen, L.; Wang, H.; Zhao, Y.; Zhang, Y.; Cumming, D.R.S.; Chen, Q. Metamaterial Absorber Integrated Micro-fluidic Terahertz Sensors. Laser Photonics Rev. 2016, 10, 962–969, doi:10.1002/LPOR.201600064.
- Zhao, Z.; Cui, H.; Song, W.; Ru, X.; Zhou, W.; Yu, X. A Simple Magnetic Nanoparticles-Based Viral RNA Extraction Method for Efficient Detection of SARS-CoV-2. bioRxiv 2020. Fischer, B.; Hoffmann, M.; Helm, H.; Modjesch, G.; Jepsen, P.U. Chemical Recognition in Terahertz Time-Domain Spectroscopy and Imaging. Semicond. Sci. Technol. 2005, 20, S246, doi:10.1088/0268-1242/20/7/015.
- Ou, H.; Lu, F.; Xu, Z.; Lin, Y.-S. Terahertz Metamaterial with Multiple Resonances for Biosensing Application. Nanomaterials 2020, 10, 1038. Yeo, W.G.; Gurel, O.; Srinivasan, N.; King, P.D.; Nahar, N.K.; Park, S.; Lehman, N.L.; Sertel, K. Terahertz Imaging and Elec-tromagnetic Model of Axon Demyelination in Alzheimer’s Disease. IEEE Transactions on Terahertz Science and Technology 2017, 7, 711–721, doi:10.1109/TTHZ.2017.2739481.
- Kim, J.; Oh, S.Y.; Shukla, S.; Hong, S.B.; Heo, N.S.; Bajpai, V.K.; Chun, H.S.; Jo, C.H.; Choi, B.G.; Huh, Y.S.; et al. Heteroassembled Gold Nanoparticles with Sandwich-Immunoassay LSPR Chip Format for Rapid and Sensitive Detection of Hepatitis B Virus Surface Antigen (HBsAg). Biosens. Bioelectron. 2018, 107, 118–122. Ahmadivand, A.; Gerislioglu, B.; Manickam, P.; Kaushik, A.; Bhansali, S.; Nair, M.; Pala, N. Rapid Detection of Infectious Envelope Proteins by Magnetoplasmonic Toroidal Metasensors. ACS Sens. 2017, 2, 1359–1368, doi:10.1021/ACSSENSORS.7B00478.
- Akter, N.; Legacy, A.; Alam, F.; Pala, N. Hybrid Toroidal Resonance Response in Planar Core-Shell THz Metasurfaces. Plas-monics 2021, 2021, 1–7, doi:10.1007/S11468-021-01427-4.
- Akter, N.; Karabiyik, M.; Pala, N. Hybrid Toroidal Modes in Planar Core-Shell Metamaterial Structures. In Proceedings of the 2018 IEEE Photonics Conference (IPC), Reston, VA, USA, 30 September–4 October 2018; pp. 1–2; doi:10.1109/IPCON.2018.8527265.
- Ahmadivand, A.; Gerislioglu, B. Large-Modulation-Depth Polarization-Sensitive Plasmonic Toroidal Terahertz Metamaterial. IEEE Photonics Technol. Lett. 2017, 29, 1860–1863, doi:10.1109/LPT.2017.2754339.
- Ahmadivand, A.; Gerislioglu, B.; Tomitaka, A.; Manickam, P.; Kaushik, A.; Bhansali, S.; Nair, M.; Pala, N. Extreme Sensitive Metasensor for Targeted Biomarkers Identification Using Colloidal Nanoparticles-Integrated Plasmonic Unit Cells. Biomed. Opt. Express 2018, 9, 373, doi:10.1364/boe.9.000373.
- Ahmadivand, A.; Gerislioglu, B.; Ahuja, R.; Kumar Mishra, Y. Terahertz Plasmonics: The Rise of Toroidal Metadevices towards Immunobiosensings. Mater. Today 2020, 32, 108–130, doi:10.1016/J.MATTOD.2019.08.002.
- Keshavarz, A.; Vafapour, Z. Sensing Avian Influenza Viruses Using Terahertz Metamaterial Reflector. IEEE Sens. J. 2019, 19, 5161–5166, doi:10.1109/JSEN.2019.2903731.
- Amin, M.; Siddiqui, O.; Abutarboush, H.; Farhat, M.; Ramzan, R. A THz Graphene Metasurface for Polarization Selective Virus Sensing. Carbon 2021, 176, 580–591, doi:10.1016/j.carbon.2021.02.051.
- Hong, J.T.; Jun, S.W.; Cha, S.H.; Park, J.Y.; Lee, S.; Shin, G.A.; Ahn, Y.H. Enhanced Sensitivity in THz Plasmonic Sensors with Silver Nanowires. Sci. Rep. 2018, 8, 15536, doi:10.1038/s41598-018-33617-2.
- Sun, Y.; Zhong, J.; Zhang, C.; Zuo, J.; Pickwell-MacPherson, E. Label-Free Detection and Characterization of the Binding of Hemagglutinin Protein and Broadly Neutralizing Monoclonal Antibodies Using Terahertz Spectroscopy. J. Biomed. Opt. 2015, 20, 037006, doi:10.1117/1.jbo.20.3.037006.
- Dolai, S.; Tabib-Azar, M. Terahertz Detection of Zika Viruses. Preprints 2020, 2020020232, doi:10.20944/preprints202002.0232.v1.
- Ahmadivand, A.; Gerislioglu, B.; Ramezani, Z.; Kaushik, A.; Manickam, P.; Ghoreishi, S.A. Functionalized Terahertz Plasmonic Metasensors: Femtomolar-Level Detection of SARS-CoV-2 Spike Proteins. Biosens. Bioelectron. 2021, 177, 112971, doi:10.1016/j.bios.2021.112971.
- Terahertz Technology for the Identification of the COVID-19. Review of Academic Literature March 2020, TERAGROUP Prepared in Cooperation with: The Israeli Ministry of Defense & Rabatit Engineering & Consulting Ltd. Available online: https://www.iclinics.cl/wp-content/uploads/2020/10/tae-Terahertz-Technology-for-the-Identification-of-the-COVID-19-Academic-Literature.pdf (accessed on 19 June 2021).
- Qiu, G.; Gai, Z.; Tao, Y.; Schmitt, J.; Kullak-Ublick, G.A.; Wang, J. Dual-Functional Plasmonic Photothermal Biosensors for Highly Accurate Severe Acute Respiratory Syndrome Coronavirus 2 Detection. ACS Nano 2020, 14, 5268–5277, doi:10.1021/acsnano.0c02439.
- Seo, G.; Lee, G.; Kim, M.J.; Baek, S.-H.; Choi, M.; Ku, K.B.; Lee, C.-S.; Jun, S.; Park, D.; Kim, H.G.; et al. Rapid Detection of COVID-19 Causative Virus (SARS-CoV-2) in Human Nasopharyngeal Swab Specimens Using Field-Effect Transistor-Based Biosensor. ACS Nano 2020, 14, 5135–5142, doi:10.1021/ACSNANO.0C02823.
- Das, C.M.; Guo, Y.; Yang, G.; Kang, L.; Xu, G.; Ho, H.P.; Yong, K.T. Gold Nanorod Assisted Enhanced Plasmonic Detection Scheme of COVID-19 SARS-CoV-2 Spike Protein. Adv. Theory Simul. 2020, 3, 2000185, doi:10.1002/adts.202000185.
- Ahmed, S.R.; Kang, S.W.; Oh, S.; Lee, J.; Neethirajan, S. Chiral Zirconium Quantum Dots: A New Class of Nanocrystals for Optical Detection of Coronavirus. Heliyon 2018, 4, e00766, doi:10.1016/j.heliyon.2018.e00766.
- Barauna, V.G.; Singh, M.N.; Barbosa, L.L.; Marcarini, W.D.; Vassallo, P.F.; Mill, J.G.; Ribeiro-Rodrigues, R.; Campos, L.C.G.; Warnke, P.H.; Martin, F.L. Ultrarapid On-Site Detection of SARS-CoV-2 Infection Using Simple ATR-FTIR Spectroscopy and an Analysis Algorithm: High Sensitivity and Specificity. Anal. Chem. 2021, 93, 2950–2958, doi:10.1021/ACS.ANALCHEM.0C04608.
- Carlomagno, C.; Bertazioli, D.; Gualerzi, A.; Picciolini, S.; Banfi, P.I.; Lax, A.; Messina, E.; Navarro, J.; Bianchi, L.; Caronni, A.; et al. COVID-19 Salivary Raman Fingerprint: Innovative Approach for the Detection of Current and Past SARS-CoV-2 Infec-tions. Sci. Rep. 2021, 11, 1–13, doi:10.1038/s41598-021-84565-3.
- Qiu, G.; Gai, Z.; Saleh, L.; Tang, J.; Gui, T.; Kullak-Ublick, G.A.; Wang, J. Thermoplasmonic-Assisted Cyclic Cleavage Ampli-fication for Self-Validating Plasmonic Detection of SARS-CoV-2. ACS Nano 2021, 15, 7536–7546, doi:10.1021/ACSNANO.1C00957.
- Funari, R.; Chu, K.Y.; Shen, A.Q. Detection of Antibodies against SARS-CoV-2 Spike Protein by Gold Nanospikes in an Opto-Microfluidic Chip. Biosens. Bioelectron. 2020, 169, 112578, doi:10.1016/J.BIOS.2020.112578.
- Zhao, Z.; Cui, H.; Song, W.; Ru, X.; Zhou, W.; Yu, X. A Simple Magnetic Nanoparticles-Based Viral RNA Extraction Method for Efficient Detection of SARS-CoV-2. bioRxiv 2020, 2020.02.22.961268, doi:10.1101/2020.02.22.961268.
- Kim, J.; Oh, S.Y.; Shukla, S.; Hong, S.B.; Heo, N.S.; Bajpai, V.K.; Chun, H.S.; Jo, C.H.; Choi, B.G.; Huh, Y.S.; et al. Heteroas-sembled Gold Nanoparticles with Sandwich-Immunoassay LSPR Chip Format for Rapid and Sensitive Detection of Hepatitis B Virus Surface Antigen (HBsAg). Biosens. Bioelectron. 2018, 107, 118–122, doi:10.1016/J.BIOS.2018.02.019.
- Deng, S.; Wang, P.; Yu, X. Phase-Sensitive Surface Plasmon Resonance Sensors: Recent Progress and Future Prospects. Sensors 2017, 17, 2819, doi:10.3390/S17122819.
- Yesilkoy, F.; Terborg, R.A.; Pello, J.; Belushkin, A.A.; Jahani, Y.; Pruneri, V.; Altug, H. Phase-Sensitive Plasmonic Biosensor Using a Portable and Large Field-of-View Interferometric Microarray Imager. Light Sci. Appl. 2018, 7, 17152, doi:10.1038/lsa.2017.152.
- Jin, B.; Tan, W.; Zhang, C.; Wu, J.; Chen, J.; Zhang, S.; Wu, P. High-Performance Terahertz Sensing at Exceptional Points in a Bilayer Structure. Adv. Theory Simul. 2018, 1, 1800070, doi:10.1002/ADTS.201800070.
- Peng, K.; Jevtics, D.; Zhang, F.; Sterzl, S.; Damry, D.A.; Rothmann, M.U.; Guilhabert, B.; Strain, M.J.; Tan, H.H.; Herz, L.M.; et al. Three-Dimensional Cross-Nanowire Networks Recover Full Terahertz State. Science 2020, 368, 510–513, doi:10.1126/SCIENCE.ABB0924.
- Sinha, R.; Karabiyik, M.; Al-Amin, C.; Vabbina, P.K.; Pala, N. Tunable Room Temperature THz Sources Based on Nonlinear Mixing in a Hybrid Optical and THz Micro-Ring Resonator. Sci. Rep. 2015, 5, 9422, doi:10.1038/srep09422.
- Cumming, D.R.S.; Liang, L.; Wen, L.; Zheng, Q.; Chen, Q.; Cumming,zik D.R.S.; Chen, Q. Miniaturized Spectroscopy with Tunable and Sensitive Plasmonic Structures. Opt. Lett. 2021, 46, 4264–4267, doi:10.1364/OL.426624.
- Liebermeister, L.; Nellen, S.; Kohlhaas, R.B.; Lauck, S.; Deumer, M.; Breuer, S.; Schell, M.; Globisch, B. Optoelectronic Fre-quency-Modulated Continuous-Wave Terahertz Spectroscopy with 4 THz Bandwidth. Nat. Commun. 2021, 12, 1–10, doi:10.1038/s41467-021-21260-x.
- Vitiello, M.S.; Consolino, L.; Inguscio, M.; Natale, P. de Toward New Frontiers for Terahertz Quantum Cascade Laser Fre-quency Combs. Nanophotonics 2021, 10, 187–194, doi:10.1515/NANOPH-2020-0429.
- Valušis, G.; Lisauskas, A.; Yuan, H.; Knap, W.; Roskos, H.G. Roadmap of Terahertz Imaging 2021. Sensors 2021, 21, 4092, doi:10.3390/S21124092.
- Son, J.H. Terahertz Bio-Sensing Techniques. In Handbook of Terahertz Technology for Imaging, Sensing and Communications; Woodhead Publishing: Sawston, UK,2013; pp. 217–230; doi:10.1533/9780857096494.2.217.
- Nagel, M.; Först, M.; Kurz, H. THz Biosensing Devices: Fundamentals and Technology. J. Phys. Condens. Matter 2006, 18, S601, doi:10.1088/0953-8984/18/18/S07.
