Your browser does not fully support modern features. Please upgrade for a smoother experience.
Please note this is a comparison between Version 1 by Pamela Dow and Version 2 by Rita Xu.
Oxycodone is a widely used opioid for the management of chronic pain. Analgesic effects observed following the administration of oxycodone are mediated mostly by agonistic effects on the μ-opioid receptor. Wide inter-subject variability observed in oxycodone efficacy could be explained by polymorphisms in the gene coding for the μ-opioid receptor (OPRM1).
- oxycodone
- pharmacogenetics
- CYP2D6
- OPRM1
- COMT
- efficacy
1. Introduction
Drug overdose deaths involving prescription opioids (natural and semi-synthetic opioids and methadone) rose from 3442 in 1999 to 14,139 in 2019, reaching a peak of 17,209 deaths in 2017 [1]. Oxycodone is a semi-synthetic opioid used for pain management, accounting for approximately 17 million prescriptions in 2018 [2]. In 2016–2017, oxycodone prescriptions represented approximately 18% of all pain management prescriptions following an emergency room visit in the United States [3]. Recently, the role of genetic polymorphisms associated with proteins involved in the pharmacokinetics and pharmacodynamics of oxycodone was reviewed by the Clinical Pharmacogenetics Implementation Consortium (CPIC), which concluded that current evidence was insufficient to make firm clinical recommendations—in the form of a clinical guideline—regarding genetic testing for oxycodone dose adjustment [4]. Some study results have clearly demonstrated and supported the value of genetic testing, though others have not found clinical value in genetic testing to improve pain management with oxycodone. This is especially the case for polymorphisms associated with CYP2D6 and the formation of oxymorphone—an active metabolite of oxycodone—and for polymorphisms linked to the opioid receptor mu 1 gene (OPMR1) [5].
2. Pharmacokinetic Considerations
2.1. CYP2D6 Activities, Polymorphisms, and Expression (GRADE High Quality ++++)
The cytochrome P450 (CYP450) superfamily is comprised of several heme-containing monooxygenases involved in the phase I metabolism of ~75% of small molecules and is a crucial part of our current pharmacological armamentarium. In humans, this superfamily of enzymes is encoded by 57 genes from 18 families [6][7]. One gene, CYP2D6, codes for an enzyme involved in the metabolism of eicosanoids, drugs, and foreign chemicals [7][8][9][10][8,9,10,11]. CYP2D6 is a highly polymorphic gene, and more than 100 variants and subvariants have been identified [11][12]. CYP2D6 substrates are typically lipophilic bases with an aromatic ring and a nitrogen atom that are protonated at physiological pH. Substrate binding is then followed by oxidation 5–7 Å, apart from the nitrogen moiety [12][13][13,14]. CYP2D6 is responsible for the clearance of at least 20% of the compounds in current clinical use, including antiarrhythmics, antidepressants, antipsychotics, β-blockers, and analgesics [8][9][14][9,10,15].
As CYP2D6 allelic variants influence protein expression and activity, CYP2D6 polymorphisms affect the functional capability of the enzyme to metabolize CYP2D6 substrate drugs. Allele combinations can produce poor metabolizers (PMs: non-functioning CYP2D6), intermediate metabolizers (IMs: low-functioning CYP2D6), normal metabolizers (NMs: normal functioning CYP2D6), and ultra-rapid metabolizers (UMs: high-functioning CYP2D6, mostly due to gene duplication). In a large genetic testing study of 104,509 deidentified patient samples, it was extrapolated that in the population, an estimated 5.7% of individuals would be PMs, 10.7% would be IMs, and 2.2% would be UMs, with 81% NMs [15][16]. However, there are differences in CYP2D6 allele frequencies in distinct racial and ethnic populations. For example, the CYP2D6*4 allele frequency appears higher in Caucasians, CYP2D6*10 in East Asians, CYP2D6*41 and duplication/multiplication of active alleles in those of Middle Eastern descent, CYP2D6*17 in Black Africans, and CYP2D6*29 in African Americans [16][17][18][17,18,19].
CYP2D6 is highly and selectively expressed in the human liver. Few tissues express CYP2D6 and recent studies demonstrated that when using an absolute protein assay and LC-MSMS technique, the protein could not be detected in various regions of the human small intestine [19][20][20,21]. In 1987, Fonne-Pfister et al. demonstrated the expression of CYP2D6 in the human brain [21][22]. More than 10 years later, dextromethorphan—a well-known CYP2D6 probe substrate—to dextrorphan metabolism was demonstrated in human brain microsomes [22][23]. CYP2D6 expression has also been described in various brain regions and cell types including the cerebral cortex, hippocampus, cerebellum, basal glia, midbrain, thalamus, and glial cells [23][24][25][26][27][24,25,26,27,28]. CYP2D6 is also involved in the metabolism of tyramine to dopamine, in the regeneration of serotonin from 5-methoxytryptamine, and in behavioral traits and psychopathology [28][29][30][31][32][29,30,31,32,33].
2.2. Oxycodone Metabolism in Humans (PK and PGx; GRADE High Quality ++++)
Drug metabolism studies have characterized the extensive biotransformation of oxycodone in humans. Oxycodone is N-demethylated to noroxycodone and O-demethylated to oxymorphone; both undergo subsequent major metabolism by either N-demethylation (noroxycodone into noroxymorphone) or glucuronidation (oxymorphone into oxymorphone 3-glucuronide) [33][34][34,35]. In vitro studies conducted with human liver microsomes and genetically-engineered recombinant drug-metabolizing enzyme products have demonstrated that CYP3A4 and CYP2D6 isoforms mediate most of oxycodone metabolism [33][35][34,36]. A small amount is converted to α/β oxycodol via 6-keto reduction and a small fraction is also directly converted to a glucuronide conjugate (Figure 1) [36][37].
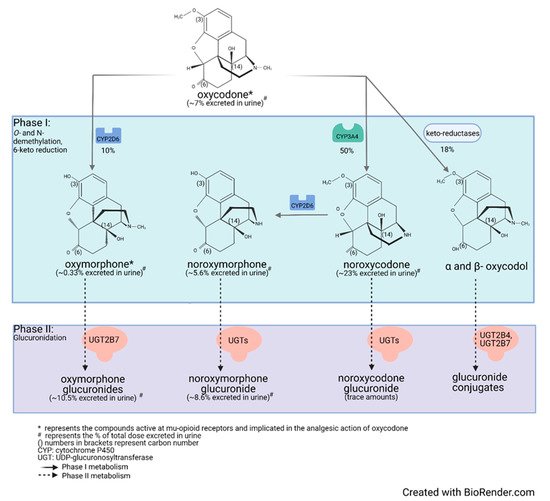

Figure 1. Detailed illustration of the elimination pathways of oxycodone.
Lalovic et al., using human liver microsomes, showed that CYP3A4 was the highest-affinity enzyme involved in the N-demethylation of oxycodone to noroxycodone with a mean Km of 600 ± 119 μM [33][34]; co-incubation with the CYP3A4 inhibitor ketoconazole decreased formation of noroxycodone by more than 90%. During the O-demethylation of oxycodone to oxymorphone, CYP2D6 was found to have the highest affinity, with a Km of 130 ± 33 μM [33][34]. Other enzymes contributed to less than 8% for the N-demethylation and 10–26% for the O-demethylation pathways, respectively. N-Demethylation activity in intestinal microsomes was 20–50% that of the liver microsomes, while the O-demethylation was negligible, suggesting that the liver may be primarily responsible for first-pass metabolism, especially for the O-demethylation [33][34]. These results are in agreement with CYP3A4 and CYP2D6 protein levels measured in the human small intestine [19][37][20,38]. Romand et al. confirmed the important roles of CYP3A4 and CYP2D6 in the phase I transformation of oxycodone, while implicating UGT2B7 (Km: 762 ± 153 μM) and to a lesser extent UGT2B4 (Km: 2454 ± 497 μM) in the phase II metabolism of oxycodone [36][37].
Human mass-balance and pharmacokinetic studies confirmed that oxycodone is extensively metabolized in humans with only 10% of the drug excreted unchanged in urine [38][39][40][39,40,41]. In agreement with in vitro studies, oral administration of oxycodone showed that CYP3A4 is involved in the major metabolic clearance pathway with about 50% of the drug being N-demethylated to noroxycodone, while about 10% is O-demethylated by CYP2D6 to oxymorphone [40][41][42][43][41,42,43,44]. Following oral administration of oxycodone (15 mg single-dose immediate-release formulation) in 10 healthy individuals, (one CYP2D6 PM, one CYP2D6 UM, eight CYP2D6 activity NMs), average peak plasma concentrations of oxycodone reached approximately 30 ng/mL, while oxymorphone peak plasma concentrations were about 0.7 ng/mL [44][45][46][47][45,46,47,48]. In plasma of CYP2D6 NMs, the oxycodone-to-oxymorphone ratio could therefore be estimated at ~43:1 (Table 1) [45][46][47][46,47,48]. Under similar conditions, noroxycodone concentrations reached about 20 ng/mL [44][45][46][47][45,46,47,48].
Table 1. Change in oxycodone:oxymorphone ratio depending on patient genotype or use of CYP450-inhibitor drugs.
| CYP2D6 Activity | Oxycodone/Oxymorphone Concentration Ratio in Plasma | Oxycodone/Oxymorphone Free-Drug Concentration Ratio in Plasma | Oxycodone/Oxymorphone Free-Drug Concentration Ratio in the Brain * | Oxycodone/Oxymorphone Relative Contribution to μ-Opioid Receptor-Binding Considering the Free-Drug Concentration Ratio in the Brain ** |
|---|
| UM CYP2D6 | 32:1 | 19:1 | 57:1 | 0.6:1 |
| NM CYP2D6 | 43:1 | 26:1 | 78:1 | 0.8:1 |
| PM CYP2D6 | 300:1 | 180:1 | 540:1 | 5.4:1 |
| With potent CYP2D6 inhibitors | 110:1 | 66:1 | 198:1 | 2:1 |
| With potent CYP3A4 inhibitors | 56:1 to 21:1 | 34:1 to 13:1 | 102:1 to 39:1 | 1:1 to 0.4:1 |
CYP2D6: cytochrome P450 2D6; UM: ultra-rapid metabolizer; NM: normal metabolizer; PM: poor metabolizer; CYP3A4: cytochrome P450 3A4; *: assuming a conservative brain-to-blood unbound concentration ratio of 3.0 for oxycodone and of 1.0 for oxymorphone; **: assuming a conservative brain-to-blood unbound concentration ratio of 3.0 for oxycodone and of 1.0 for oxymorphone and a potency ratio of 1:100 for oxycodone vs. oxymorphone.
Valuable information on the relative contributions of CYP2D6 and CYP3As to the disposition of oxycodone can also be obtained from pharmacogenomic and drug–drug interactions studies. Studies performed in individuals with multiple copies of CYP2D6—which translates to an UM phenotype—have shown that average plasma levels of oxycodone are reduced while average concentrations of oxymorphone are increased (relative to NMs). The oxycodone-to-oxymorphone plasma-concentration ratio was then estimated to be ~32:1 in UMs following oral administration (Table 1) [45][46][47][46,47,48].
In patients and healthy volunteers with non-functional CYP2D6 (PMs), the oxycodone-to-oxymorphone ratio is increased to approximately 300:1 (Table 1) [47][48][48,49]. This increased ratio is mostly explained by a significant decrease in oxymorphone plasma concentrations, as the overall contribution of CYP2D6 to the total clearance of oxycodone is limited. Still, a nominal amount of oxymorphone is detected in the plasma of these individuals, suggesting that an isoform other than CYP2D6 could also mediate the formation of oxymorphone. Similar results were obtained during in vitro drug metabolism studies [33][34]. Notably, much lower ratios of oxycodone/oxymorphone were observed in patients from all phenotypic groups following the administration of oxycodone intravenously [49][50]. This observation can be explained by a route of administration-dependent difference in the relative contributions of CYP3A4 and CYP2D6 to the clearance of oxycodone, as CYP2D6 expression is limited in the intestinal tissue [20][50][51][52][21,51,52,53]. Hence, the relative contribution of CYP2D6 to the total clearance of oxycodone appears increased following intravenous administration.
Drug interactions studies have been conducted with potent CYP2D6 inhibitors. Heiskanen et al. and Sirhan-Daneau et al. conducted studies in normal CYP2D6 metabolizers and demonstrated—using quinidine as a non-competitive and potent inhibitor of CYP2D6—that formation of oxymorphone was almost completely impeded under potent CYP2D6 inhibition [45][46][53][54][46,47,54,55]. However, using sensitive LC-MS/MS assays, limited amounts of oxymorphone could still be detected in their plasma [53][54]. Similar results were obtained by Lemberg et al. using paroxetine as a mechanism-based inhibitor of CYP2D6 [55][56]. Under potent inhibition of CYP2D6, the oxycodone-to-oxymorphone plasma-concentration ratio is nearly ~110:1 (Table 1).
As mentioned previously, CYP3A4 is the major enzyme involved in the disposition of oxycodone. Inhibition of CYP3A4, using inhibitory agents such as itraconazole or ketoconazole, is associated with increases in both oxycodone and also oxymorphone levels. As CYP3A4 is inhibited, more oxycodone is available for its metabolism by CYP2D6 into oxymorphone. Under conditions of CYP3A4 inhibition, the oxycodone to oxymorphone plasma concentration ratios ranged from 56:1 to 21:1 (Table 1) [56][57][58][57,58,59].
2.3. Oxycodone Distribution and Protein Binding (PK; GRADE Low Quality ++−−)
Average plasma protein binding for oxycodone is 45%, while that for oxymorphone is 11%. Hence, for all situations described above, the relative oxycodone/oxymorphone ratios while considering free-drug concentrations in the plasma [(Cavg oxycodone × Fu)/(Cavg oxymorphone × Fu)] should be decreased by about 40% (Table 1).
Drug distribution studies have demonstrated that both oxycodone and oxymorphone can cross the blood–brain barrier. First, Bostrom et al. reported in two different studies that the brain-to-blood unbound concentration ratio of oxycodone was 3.0–6.0 [59][60][60,61]. Then, Zasshi et al. also reported that the brain-to-blood unbound concentration ratio for oxycodone was 3.0 [61][62]. In regards to oxymorphone, Sadiq et al. reported that the brain-to-blood unbound concentration ratio for oxymorphone was 1.9 [62][63]. In contrast, Zasshi et al. and Lalovic et al. reported that the oxymorphone brain-to-blood unbound concentration ratio was 0.26 to 0.3 [33][43][61][34,44,62]. However, the discrepancy between these results could not be explained. Assuming a conservative brain-to-blood unbound concentration ratio of 3.0 for oxycodone and an average of 1.0 for oxymorphone, new oxycodone-to-oxymorphone ratios can be estimated (Table 1); noroxycodone and noroxymorphone do not have good brain penetration [43][44].
2.4. Oxymorphone Pharmacokinetics Following Direct Oxymorphone Administration (PK; GRADE Moderate Quality +++−)
Oxymorphone is a known potent μ-opioid receptor agonist which may contribute, totally or partially, to the overall analgesic effects observed following oxycodone oral administration [63][64]. Oxymorphone can be administered directly as an active drug to humans and its pharmacokinetics and pharmacodynamics can be appreciated and compared to its pharmacokinetics/pharmacodynamic relationship observed following oxycodone administration.
The bioavailability of oral oxymorphone is about 10% following an important first-pass metabolism. Oxymorphone undergoes extensive conjugation mediated mostly by UGT2B7 to produce its primary metabolite, oxymorphone 3-glucuronide [64][65][65,66]. To a lesser extent, it also undergoes reduction of the 6-ketone group to form 6-hydroxy-oxymorphone. After administration of a single 10 mg oxymorphone immediate-release oral dose, 1.9% was eliminated in urine as free oxymorphone, 44.1% as conjugated oxymorphone 3-glucuronide, 0.3% as 6-hydroxy-oxymorphone, and 2.6% as conjugated 6-hydroxy-oxymorphone [34][35]. Note that in contrast to other opioids, the N-demethylation of oxymorphone into noroxymorphone is very limited in humans [34][35].
Following the oral administration of a single extended-release 10 mg dose of oxymorphone, peak plasma concentrations of oxymorphone, 6-hydroxy-oxymorphone, and oxymorphone 3-glucuronide reached 0.65 ng/mL, 0.37 ng/mL, and 112 ng/mL, respectively [66][67]. Plasma levels of oxymorphone observed under these conditions were similar to plasma concentrations observed following administration of a single 15 mg immediate-release oxycodone dose (0.7 ng/mL) [45][46][54][46,47,55]. This observation strongly suggests that oxymorphone plasma levels of 0.5 to 1.0 ng/mL mediate clinically relevant analgesic effects and likely contribute significantly to oxycodone efficacy.
