Your browser does not fully support modern features. Please upgrade for a smoother experience.
Please note this is a comparison between Version 2 by Rita Xu and Version 1 by Carla Pires.
COVID-2019 pandemic lead to a raised interest on the development of new treatments through Artificial Intelligence (AI). AI is a suitable tool to quickly analyze large amounts of data or to estimate drug repurposing against COVID-2019.
- artificial intelligence
- COVID-2019
- SARS-CoV-2
- drug design
- medicines
- repurposing of drugs
- molecular docking
1. Introduction
On 31 December 2019, a cluster of cases of pneumonia in Wuhan, Hubei Province, China was reported with the identification of a likely new form of coronavirus. The genetic sequence of the virus SARS-CoV-2 (severe acute respiratory syndrome coronavirus 2), which is the etiologic agent of COVID-2019 (coronavirus disease 2019) was shared by China on 12 January 2020. The first COVID-2019 case was reported outside China in Thailand on 13 January 2020, which was followed by an alarming spread of the number of cases in different regions/countries. On 11 March 2020, the COVID-2019 pandemic was declared by the World Health Organization (WHO) [1,2][1][2]. Globally, 188,655,968 confirmed cases of COVID-19, including 4,067,517 deaths have been reported to WHO on 16 July 2021, with 3,402,275,866 vaccine administered doses on 15 July 2021 [3].
SARS-CoV-2 infects the epithelial cells or immune cells, producing tissue damage. Epithelial cells and immune cells release inflammatory cytokines, such as IL-1, IL-6, IL-12, and TNFα. Inflammatory cytokines recruit innate immune cells (e.g., monocytes, macrophages, neutrophils, DCs, and NK cells) and activate adaptive immune cells (CD4+ T cells and CD8+ T cells). These occurrences may induce myelopoiesis and emergency granulopoiesis, associated with the production of sustained and excessive circulating cytokines, which may lead to a serious epithelial damage. Immunopathologic events may contribute to explain COVID-19 pathogenesis, such as lymphopenia, neutrophilia, dysregulation of monocytes and macrophages, reduced/delayed response of type I interferon (IFN-I), antibody-dependent enhancement, and cytokine storm [4]. The knowledge of COVID-2019 immunopathology may help in the comprehension and development of new treatments and/or drug repurposing (i.e., the application of old drugs to treat new diseases) [5,6,7,8][5][6][7][8]. Additionally, the measurement of serum levels of pro-inflammatory cytokines (e.g., IL-6) may be applied in the management of COVID-2019 (adult and pediatric patients), such as in the assessment of risk, monitoring of disease progression, determination of prognosis, selection of therapy. and prediction of response to treatment [7].
Because of the pandemic, the research on the development of new therapies, treatments, and vaccines against COVID-2019, including drug repurposing assumed an unprecedent significance at a global level [5]. Drug repurposing present some advantages over the discovery of new molecules/active substances, such as the reduction of production costs, the achievement of likely safe medicines, and potentially shorter development timelines [5,6][5][6].
The development of new treatments against COVID-2019 keep on a hot topic due to the likely risk of new virus variants, which may compromise the efficacy of treatments [9]. By definition, a “variant has one or more mutations that differentiate it from other variants in circulation” [10]. These variants may comprise the efficacy of approved treatments for COVID-2019, such as vaccines and/or other medicines. Additionally, the number of therapeutics against COVID-2019 remains limited [9,11][9][11].
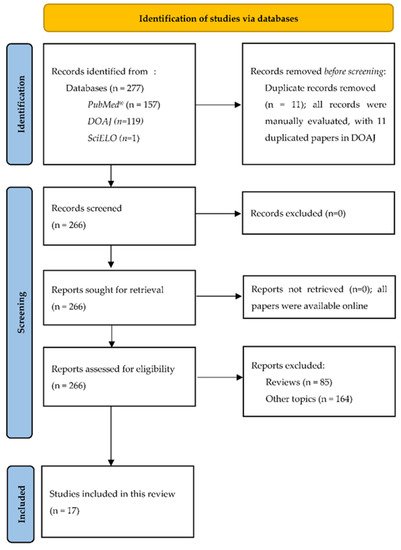
2. PRISMA Flow Diagram
The Systematic Reviews and Meta-Analyses (PRISMA) flow diagram is presented in Figure 1 [27][12].
3. Main Findings of the Selected Studies
Some aspects of the 17 selected studies are briefly summarized in Table 1, such as the list of key repurposed drug(s) with potential therapeutic use against COVID-2019.
Table 1.
Some relevant aspects of the 17 selected studies.
| Author(s); Year Reference |
Country(ies) of Origin | Key Repurposed Drug(s) Potentially Relevant against the Treatment of COVID-2019 | Key Used AI Methodologies |
|---|
| (Ke, Peng, Yeh, et al., 2020) [28] | Taiwan | Bedaquiline Brequinar Celecoxib Clofazimine Conivaptan Gemcitabine Tolcapone Vismodegib |
First, an an AI platform was defined to identify potential old/repurposed drugs with anti-coronavirus activities (or potential anti-coronavirus activity). Second, AI predicted drugs were tested for their activity against a feline coronavirus in vitro. Third, results of assays were introduced in the AI system. A Deep Neural Network algorithm was used to identify the most relevant descriptors, with the generation of different weightings to generate AI prediction models. |
| (Ke, Peng, Yeh, et al., 2020) [13] | Taiwan | Bedaquiline Brequinar Celecoxib Clofazimine Conivaptan Gemcitabine Tolcapone Vismodegib |
First, an an AI platform was defined to identify potential old/repurposed drugs with anti-coronavirus activities (or potential anti-coronavirus activity). Second, AI predicted drugs were tested for their activity against a feline coronavirus in vitro. Third, results of assays were introduced in the AI system. A Deep Neural Network algorithm was used to identify the most relevant descriptors, with the generation of different weightings to generate AI prediction models. |
| (Morselli, do Valle, Zitnik, et al., 2021) [29] | USA, Turkey, Hungary | Auranofin | AI based on algorithms, network diffusion, and network proximity, tasking to rank 6,340 drugs, regarding their potential efficacy against SARS-CoV-2. Multimodal technology was required to fuse the prediction of all algorithms, since the predictive algorithm did not offer consistently reliable outcomes. The top-ranked drugs were screened in human cells. |
| (Morselli, do Valle, Zitnik, et al., 2021) [14] | |||
| Azelastine | Fluvastatin Methotrexate | ||
| USA, Turkey, Hungary | |||
| Vinblastine | |||
| Auranofin | Azelastine Fluvastatin Methotrexate Vinblastine |
AI based on algorithms, network diffusion, and network proximity, tasking to rank 6,340 drugs, regarding their potential efficacy against SARS-CoV-2. Multimodal technology was required to fuse the prediction of all algorithms, since the predictive algorithm did not offer consistently reliable outcomes. The top-ranked drugs were screened in human cells. | |
| (Abdulla, Wang, Qian, et al., 2020) [30] * | China, Singapore | Amantadine Azithromycin Chloroquine Omeprazole Sodium (optional) Ribavirin (optional) - These repurposing drugs were evaluated in combination. |
An AI-based platform was used to interrogate 12 drug/dose parameters space. Combination therapies that optimally inhibit A549 lung cell infection by vesicular stomatitis virus within 3 days of project start were identified. This AI project utilized a quadratic relationship between drug/dose inputs and efficacy/safety outputs to successfully identify the drug-dose parameter space. |
| (Abdulla, Wang, Qian, et al., 2020) [15] * | China, Singapore | Amantadine Azithromycin Chloroquine Omeprazole Sodium (optional) Ribavirin (optional) - These repurposing drugs were evaluated in combination. |
An AI-based platform was used to interrogate 12 drug/dose parameters space. Combination therapies that optimally inhibit A549 lung cell infection by vesicular stomatitis virus within 3 days of project start were identified. This AI project utilized a quadratic relationship between drug/dose inputs and efficacy/safety outputs to successfully identify the drug-dose parameter space. |
| (Blasiak, Lim, Seah, et al., 2020) [31] * | China, Singapore, USA | Lopinavir Remdesivir Ritonavir - These repurposing drugs were evaluated in combination. |
A platform (IDentif.AI) that pairs experimental validation with AI and digital drug development was used. Workflow of the project IDentif.AI: 1) clinically relevant concentrations based on and dose–response curves and maximal plasma concentration; 2) in vitro testing of combination therapies; combination therapies were determined through an orthogonal array composite (OACD) design; 3) IDentif.AI analysis: drug–drug interactions and clinically relevant drug-dosage combinations; and 4) biological validation. |
| (Blasiak, Lim, Seah, et al., 2020) [16] * | China, Singapore, USA | Lopinavir Remdesivir Ritonavir - These repurposing drugs were evaluated in combination. |
A platform (IDentif.AI) that pairs experimental validation with AI and digital drug development was used. Workflow of the project IDentif.AI: 1) clinically relevant concentrations based on and dose–response curves and maximal plasma concentration; 2) in vitro testing of combination therapies; combination therapies were determined through an orthogonal array composite (OACD) design; 3) IDentif.AI analysis: drug–drug interactions and clinically relevant drug-dosage combinations; and 4) biological validation. |
| Studies with confirmatory in-vitro and/or clinical data (Section 3.2.2) | |||
| Studies with confirmatory in-vitro and/or clinical data | |||
| (Schultz, Vera, Sinclair et al., 2020) [32] | USA | Baricitinib | |
| (Schultz, Vera, Sinclair et al., 2020) [17] | USA | Baricitinib | |
| (Stebbing, Krishnan, de Bono et al., 2020) [33] | UK, USA, Italy, Sweden, Singapore | Baricitinib | BenevolentAI (an artificial AI platform) identified baricitinib as a potential COVID-19 drugs. Details/information on BenevolentAI works were limited. |
| (Stebbing, Krishnan, de Bono et al., 2020) [18] | UK, USA, Italy, Sweden, Singapore | Baricitinib | BenevolentAI (an artificial AI platform) identified baricitinib as a potential COVID-19 drugs. Details/information on BenevolentAI works were limited. |
| Repurposing of drugs against COVID-2019 (Section 3.2.3) | |||
| Repurposing of drugs against COVID-2019 | |||
| (Nayarisseri, Khandelwal, Madhavi et al., 2020) [34] | India, Saudi Arabia | Aprepitant Fulvestrant Remdesivir Valrubicin |
A machine learning approach was employed. Particularly, repurposed drugs were selected based on their capacity of targeting the main coronavirus protease (6LU7) using ligand-receptor Docking (optimized potential for liquid simulations algorithms to identify high affinity compounds). Additionally, candidates were subjected to Molecular Dynamic Simulations followed by ADMET (absorption, distribution, metabolism, excretion, and toxicity) studies. |
| (Nayarisseri, Khandelwal, Madhavi et al., 2020) [19] | India, Saudi Arabia | Aprepitant Fulvestrant Remdesivir Valrubicin |
A machine learning approach was employed. Particularly, repurposed drugs were selected based on their capacity of targeting the main coronavirus protease (6LU7) using ligand-receptor Docking (optimized potential for liquid simulations algorithms to identify high affinity compounds). Additionally, candidates were subjected to Molecular Dynamic Simulations followed by ADMET (absorption, distribution, metabolism, excretion, and toxicity) studies. |
| (Kim, Zhang, Cha et al., 2020) [35] | USA | Emricasan Fosamprenavir Glutamine Glutathione Piperacillin sodium Ruxolitinib Vitamin E |
Two computational approaches were applied. Fist, a high-throughput AI-based binding affinity prediction platform was used to identify FDA approved drugs with potential capacity to block coronaviruses from entering cells by binding to ACE2 (angiotensin-converting enzyme) or TMPRSS2 (Transmembrane Serine Protease 2). Second, the Disease Cancelling Technology (DCT) platform was used to identify FDA approved drugs, which may attenuate the gene expression patterns induced by coronaviruses. |
| (Kim, Zhang, Cha et al., 2020) [20] | USA | Emricasan Fosamprenavir Glutamine Glutathione Piperacillin sodium Ruxolitinib Vitamin E |
Two computational approaches were applied. Fist, a high-throughput AI-based binding affinity prediction platform was used to identify FDA approved drugs with potential capacity to block coronaviruses from entering cells by binding to ACE2 (angiotensin-converting enzyme) or TMPRSS2 (Transmembrane Serine Protease 2). Second, the Disease Cancelling Technology (DCT) platform was used to identify FDA approved drugs, which may attenuate the gene expression patterns induced by coronaviruses. |
| (Das G., Das, T., Chowdhury et al., 2021) [36] | India | Ascorbyl palmitrate Cinametic acid Guaifenesin Lauric acid Nabumetone Nafcillin Octacosanol Palmidrol Salmeterol |
|
| India | Ascorbyl palmitrate Cinametic acid Guaifenesin Lauric acid | ||
| AI deep learning techniques, in silico methodologies) and pattern recognition techniques were used to screen FDA approved pharmaceuticals and nutraceuticals to target CoV envelope (E) protein. A protein involved in the assembly and release of the virus inside the host. Multiple opensource drug databases were considered, such as ChEMBL v.26, Enamine Bio reference Compounds ( | https://www.enaminestore.com/products/bioreference-compounds | , accessed on 17 September 2021) and Chemoinformatic tools and database ( | https://chemoinfo.ipmc.cnrs.fr/TMP/tmp.32396/e-Drug3D_1930_v3.sdf, accessed on 17 September 2021). |
| (Das G., Das, T., Chowdhury et al., 2021) [21] | Nabumetone Nafcillin Octacosanol Palmidrol Salmeterol |
AI deep learning techniques, in silico methodologies) and pattern recognition techniques were used to screen FDA approved pharmaceuticals and nutraceuticals to target CoV envelope (E) protein. A protein involved in the assembly and release of the virus inside the host. Multiple opensource drug databases were considered, such as ChEMBL v.26, Enamine Bio reference Compounds (https://www.enaminestore.com/products/bioreference-compounds, accessed on 17 September 2021) and Chemoinformatic tools and database (https://chemoinfo.ipmc.cnrs.fr/TMP/tmp.32396/e-Drug3D_1930_v3.sdf, accessed on 17 September 2021). | |
| (Rajput, Thakur, Mukhopadhyay et al., 2021) [37] | India | Alatrofloxacin Metergoline Rescinnamine Rescinnamine Telotristat ethyl Verteporfin |
Robust computational methods using machine learning techniques, such as Support Vector Machine, Random Forest, k-Nearest Neighbour, Artificial Neural Network, and Deep Learning were developed by the authors to predict the repurposed drugs. |
| (Rajput, Thakur, Mukhopadhyay et al., 2021) [22] | India | Alatrofloxacin Metergoline Rescinnamine Rescinnamine Telotristat ethyl Verteporfin |
Robust computational methods using machine learning techniques, such as Support Vector Machine, Random Forest, k-Nearest Neighbour, Artificial Neural Network, and Deep Learning were developed by the authors to predict the repurposed drugs. |
| (Li, Yao, Cheng et al., 2021) [38] | China, USA | Baricitinib Bivalirudin Fostamatinib Lusutrombopag Simvastatin |
1) Public genetic screening data were successively interrogated to identify human-specific host dependency genes, i.e., indispensable genes for effective viral infections. 2) Extensive drug-target interactions were interrogated through diverse methodologies, such as database retrieval, literature mining and de novo prediction using AI-based algorithms. |
| (Li, Yao, Cheng et al., 2021) [23] | China, USA | Baricitinib Bivalirudin Fostamatinib Lusutrombopag Simvastatin |
1) Public genetic screening data were successively interrogated to identify human-specific host dependency genes, i.e., indispensable genes for effective viral infections. 2) Extensive drug-target interactions were interrogated through diverse methodologies, such as database retrieval, literature mining and de novo prediction using AI-based algorithms. |
| (McCoy, Gudapati, He, Horlander, 2021) [39] | USA | ||
| (McCoy, Gudapati, He, Horlander, 2021) [24] | |||
| Amprenavir | Albuterol Artemisinin Chloroquine Ciprofloxacin Cyclosporine Fluoroquinolones Hydroxymethylglutaryl-CoA reductase inhibitors Methotrexate Quinolone antibacterial agents Suramin Zidovudine |
||
| USA | Amprenavir Albuterol Artemisinin Chloroquine Ciprofloxacin Cyclosporine Fluoroquinolones Hydroxymethylglutaryl-CoA reductase inhibitors Methotrexate Quinolone antibacterial agents Suramin Zidovudine | ||
| A link prediction model was developed (an AI text mining model). The biomedical knowledge graph, SemNe was used to predict missing links in biomedical literature, regarding drug repurposing. TransE, CompleX, and RotatE based methods were used to visualize knowledge graph embeddings and link prediction results using in a web application. | |||
| A link prediction model was developed (an AI text mining model). The biomedical knowledge graph, SemNe was used to predict missing links in biomedical literature, regarding drug repurposing. TransE, CompleX, and RotatE based methods were used to visualize knowledge graph embeddings and link prediction results using in a web application. | |||
| (Chakravarty, Antontsev, Khotimchenko et al., 2021) [40] | USA | Captopril Lisinopril Spirapril |
The plataform BIOiSIM (an AI-integrated mechanistic modeling platform) was used to simulate systemic therapy of Calcium Channel Blockers (CCBs) and ACE compounds in tissues related to the COVID-19 pathogenesis, namely the disposition and site-of-action penetration (in silico modeling). |
| (Chakravarty, Antontsev, Khotimchenko et al., 2021) [25] | USA | Captopril Lisinopril Spirapril |
The plataform BIOiSIM (an AI-integrated mechanistic modeling platform) was used to simulate systemic therapy of Calcium Channel Blockers (CCBs) and ACE compounds in tissues related to the COVID-19 pathogenesis, namely the disposition and site-of-action penetration (in silico modeling). |
| BIOiSIM is a | |||
| dynamic, biology-driven platform that provides a scalable computational prediction of in vivo pharmacokinetic-pharmacodynamic (PK-PD) phenomena. | |||
| (Kadioglu, Saeed & Efferth, 2021) [41] | Germany | Conivaptan Dihydroergotamine Eltrombopag Ergotamine Eribulin Idarubicin Ivermectin Paritaprevir |
Diverse combined in silico methods (virtual drug screening, molecular docking, and supervised machine learning algorithms) were used in a workflow to identify repurposed drug against COVID-19. |
| (Kadioglu, Saeed & Efferth, 2021) [26] | |||
| Ledipasvir | Lifitegrast Lumacaftor Nilotinib Nystatin Ponatinib Regorafenib Rifapentine | ||
| Germany | |||
| Simeprevir | Teniposide Trabectedin Trypan blue Velpatasvir Venetoclax | ||
| Conivaptan | Dihydroergotamine Eltrombopag Ergotamine Eribulin Idarubicin Ivermectin Ledipasvir Lifitegrast Lumacaftor Nilotinib Nystatin Paritaprevir Ponatinib Regorafenib Rifapentine Simeprevir Teniposide Trabectedin Trypan blue Velpatasvir Venetoclax |
Diverse combined in silico methods (virtual drug screening, molecular docking, and supervised machine learning algorithms) were used in a workflow to identify repurposed drug against COVID-19. | |
| Repurposing of drugs against COVID-2019: association of drugs (Section 3.2.3.1) | |||
| Repurposing of drugs against COVID-2019: association of drugs | |||
| (Artigas, Coma, Matos-Filipe et al., 2020) [42] | Spain | Pirfenidone plus melatonin | The mechanism of action of pirfenidone and melatonin was simulated by using the previously described Therapeutic Performance Mapping System (TPMS) technology (an AI-based approach). GUILDify v2.0 web server was used to confirm the effect of pirfenidone and melatonin against SARS-CoV-2 infection. This web server is able to calculate the neighbourhoods of the human biological network related to the host-key points (e.g., for SARS-CoV infection) and simultaneously affected by specific drugs. |
| (Artigas, Coma, Matos-Filipe et al., 2020) [27] | Spain | Pirfenidone plus melatonin | The mechanism of action of pirfenidone and melatonin was simulated by using the previously described Therapeutic Performance Mapping System (TPMS) technology (an AI-based approach). GUILDify v2.0 web server was used to confirm the effect of pirfenidone and melatonin against SARS-CoV-2 infection. This web server is able to calculate the neighbourhoods of the human biological network related to the host-key points (e.g., for SARS-CoV infection) and simultaneously affected by specific drugs. |
| Note: References [30,31] are also related to the combination of repurposing medicines. | |||
| Note: References [15][16] are also related to the combination of repurposing medicines. | |||
| Repurposing of drugs: alternative therapies (Section 3.2.3.2) | |||
| (Wang, Li; Song, et al., 2021) [43] | China, Australia | Zhongqifangzi (PMSP) Recommended as supplementary treatment against COVID-2019. |
An ontology-based side-effect prediction framework (OSPF) was developed based on a previous work and Artificial Neural Network (ANN)-based deep learning. The Traditional Chinese Medicine prescriptions for the treatment of COVID-19 (officially recommended in China) were evaluated. |
| (Wang, Li; Song, et al., 2021) [28] | |||
| GCT-CJ | Hanshiyufen fang (HSYF-F) Huashi Baidu Formula (HSBD-F) Qingfei Paidu Decoction (QFPD-T) Shenfu zhusheye (SF-ZSY) | ||
| China, Australia | |||
| GCT-CJ | Hanshiyufen fang (HSYF-F) Huashi Baidu Formula (HSBD-F) Qingfei Paidu Decoction (QFPD-T) Shenfu zhusheye (SF-ZSY) Zhongqifangzi (PMSP) Recommended as supplementary treatment against COVID-2019. |
An ontology-based side-effect prediction framework (OSPF) was developed based on a previous work and Artificial Neural Network (ANN)-based deep learning. The Traditional Chinese Medicine prescriptions for the treatment of COVID-19 (officially recommended in China) were evaluated. | |
| (Li, Yao, Cheng et al., 2021) [38] | China, USA | Atropine (Lycii Cortex, Hyoscyami Semen) Dehydroeffusal (Junci Medulla) Lysergol (Pharbitidis Semen) Solanocapsine (Solanum Nigrum) Solanocapsine (Solanum Nigrum) Vitexifolin C (Viticis Fructus) |
|
| Atropine (Lycii Cortex, Hyoscyami Semen) | Dehydroeffusal (Junci Medulla) Lysergol (Pharbitidis Semen) Solanocapsine (Solanum Nigrum) Solanocapsine (Solanum Nigrum) | ||
| 1) Public genetic screening data were successively interrogated to identify human-specific host dependency genes, i.e., indispensable genes for effective viral infections. 2) Extensive drug-target interactions were interrogated through diverse methodologies, such as database retrieval, literature mining and de novo prediction using AI-based algorithms. | |||
| (Li, Yao, Cheng et al., 2021) [23] | China, USA | Vitexifolin C (Viticis Fructus) | 1) Public genetic screening data were successively interrogated to identify human-specific host dependency genes, i.e., indispensable genes for effective viral infections. 2) Extensive drug-target interactions were interrogated through diverse methodologies, such as database retrieval, literature mining and de novo prediction using AI-based algorithms. |
| (Kadioglu, Saeed & Efferth, 2021) [41] | Germany | Amyrin Baicalin Crinine Euphol Forsythiaside Friedelin Hoslunddiol IlexsaponinB1 IlexsaponinB2 IlexsaponinB3 Loniflavone Procyanidin Punicalagin Quercetin Quercetin-3-o-rutinoside Rutin Strictinin TirucallinA TingeninB Wogonoside ZINC252515584 ZINC27215482 ZINC15675938 |
Diverse combined in silico methods (virtual drug screening, molecular docking, and supervised machine learning algorithms) were used in a workflow to identify repurposed drug against COVID-19. |
| (Kadioglu, Saeed & Efferth, 2021) [26] | Germany | Amyrin Baicalin Crinine Euphol Forsythiaside Friedelin Hoslunddiol IlexsaponinB1 IlexsaponinB2 IlexsaponinB3 Loniflavone Procyanidin Punicalagin Quercetin Quercetin-3-o-rutinoside Rutin Strictinin TirucallinA TingeninB Wogonoside ZINC252515584 ZINC27215482 ZINC15675938 |
Diverse combined in silico methods (virtual drug screening, molecular docking, and supervised machine learning algorithms) were used in a workflow to identify repurposed drug against COVID-19. |
| (Acharya, Agarwal, Baker et al., 2020) [44] | |||
| (Acharya, Agarwal, Baker et al., 2020) [ | |||
| USA, Italy | |||
| 29] | |||
| Hypericin | Quercetin | An enhanced sampling molecular dynamics (MD) and ensemble docking was used supercomputer-driven pipeline for in silico drug discovery. | |
| USA, Italy | Hypericin Quercetin |
An enhanced sampling molecular dynamics (MD) and ensemble docking was used supercomputer-driven pipeline for in silico drug discovery. | |
* Studies also reported in the section on repurposing of drugs against COVID-2019: association of drugs (Section 3.2.3.1).
These studies are briefly described in three sections: studies with confirmatory in-vitro data (Section 3.2.1); studies with confirmatory in-vitro and/or clinical data (Section 3.2.2) and repurposing of drugs against COVID-2019 (Section 3.2.3).
3.1. Studies with Confirmatory In-Vitro Data
Bedaquiline (used to treat tuberculosis), brequinar (immunosuppressant), celecoxib (nonsteroidal anti-inflammatory drug), clofazimine (bactericidal effect on Mycobacterium leprae), conivaptan (nonpeptide, dual antagonist of arginine vasopressin; treatment of euvolemic or hypervolemic hyponatremia), gemcitabine (antineoplastic anti-metabolite), tolcapone (catechol-O-methyltransferase inhibitor used in the therapy of Parkinson disease), and vismodegib (selectively binds to and inhibits the transmembrane protein Smoothened homologue; antineoplastic activity) were identified through AI learning and prediction processes. These 8 predicted drugs were tested for activities against a feline coronavirus in an in vitro cell-based assay. The antiviral effects were measured in feline infectious peritonitis (FIP) virus-infected feline catus whole fetus-4 (Fcwf-4) cells. All predicted drugs exhibited in vitro activities against the proliferation of a FIP virus in Fcwf-4 cells for verification of antiviral activity. Two learning database collections were applied in AI searching: one from approved drugs for SARSCoV-2, HIV, influenza, and others and other from 3CL protease inhibitors. The AI search was based on the characteristics of three types of molecular descriptors, i.e., drugs with certain molecular characteristics were preferably selected [28][13].
Overall, 6340 drugs were screened for their expected efficacy against SARS-CoV-2 through algorithms based on artificial intelligence, network diffusion, and network proximity. The predicted candidates were compared to 918 compounds that had been experimentally screened for their efficacy against SARS-CoV-2 in VeroE6 cells, kidney epithelial cells derived from African green monkey. VeroE6 cells were preincubated with the drugs (8 μM–8 nM), which were challenged with wild-type SARS-CoV-2 strain USA-WA1/2020. Additionally, the list of drugs in clinical trials with potential COVID-19 efficacy was screened. Overall, 13 candidates registered positive outcomes in VeroE6 cells, which were tested in human cells to confirm their clinical relevance (Huh7 cells, in a nine-point dilution series from 25 μM–100 nM). Among the most promising candidates, were Auranofin (antirheumatic used to treat active, progressive, or destructive forms of inflammatory arthritis; DrugBank), azelastine (histamine H1-receptor antagonist used intranasally to treat allergic and vasomotor rhinitis and in an ophthalmic solution to treat allergic conjunctivitis; DrugBank), digoxin (cardiac glycoside used in the treatment of mild to moderate heart failure and for ventricular response rate control in chronic atrial fibrillation; DrugBank), and vinblastine (vinca alkaloid used to treat breast cancer, testicular cancer, neuroblastoma, Hodgkin’s and non-Hodgkins lymphoma, mycosis fungoides, histiocytosis, and Kaposi’s sarcoma; DrugBank) presented a very strong anti–SARS-CoV-2 response in human cells; fluvastatin (an HMG-CoA reductase inhibitor used to lower lipid levels and reduce the risk of cardiovascular disease including myocardial infarction and stroke; DrugBank) presented a weaker response; and methotrexate was effective, but only at the highest concentration [29][14].
An AI-based platform was applied to identify combination therapies that optimally inhibited the infection of the A549 lung cell line by vesicular stomatitis virus (efficacy) while maximizing A549 viability. This technology may be applied to address a broad spectrum of infectious diseases, including the appearance of new strains e.g., COVID-2019. For instance, Amantadine (used to treat dyskinesia in Parkinson’s patients receiving levodopa, as well as extrapyramidal side effects of medications; DrugBank); Azithromycin (macrolide antibiotic used to treat a variety of bacterial infections; DrugBank); Chloroquine (antimalarial drug, see Appendix A); Omeprazole Sodium (a proton pump inhibitor used to treat GERD associated conditions such as heartburn and gastric acid hypersecretion, and to promote healing of tissue damage and ulcers caused by gastric acid and H. pylori infection; DrugBank) and/or Ribavirin (a guanosine nucleoside used to treat some forms of Hepatitis C; DrugBank) in combination with other candidates were among the AI-optimized regimes. Optimally and sub-optimally dosed combinations, registered a sevenfold difference in efficacy, which clearly proves the critical relevance of ideal drug and dose selection/identification [30][15].
IDentif.AI is a platform that combines AI, digital drug development and in-vitro experimentation (SARS-CoV-2 in vitro, cellular infectious disease model) to detect drug interactions and optimize the design of associations therapies with suitable doses. Viral inhibition and cell cytotoxicity of drug monotherapies and drug combinations were carried out with Vero E6 cells (African green monkey kidney). The Vero E6 cells (2 × 104 cells/well) and media with and without SARS-CoV-2 treatment (100 TCID50) were added to the plates containing the drugs and the controls. The viral infection model was based on virus’s cytopathic effect (CPE). The Viral ToxGlo assay quantifies viral-induced CPEs in host cells by using cellular ATP as a marker: the decrease in cellular ATP detected is proportional to the number of viable host cells in culture, i.e., it is possible to correlate viral CPE with viral burden. The optimized combination of remdesivir (nucleoside analog), ritonavir (HIV protease inhibitor), and lopinavir (HIV-1 protease inhibitor) was identified as potentially clinically relevant. In experimental studies, a 6.5-fold enhanced efficacy in comparison to remdesivir alone. In opposition, the combination of hydroxychloroquine (antimalarial) and azithromycin (macrolide antibiotic) were relatively ineffective in vitro at clinically relevant doses [31][16]. For more details on the pharmacologic activity of remdesivir, ritonavir, lopinavir, hydroxychloroquine and azithromycin see Appendix A [19][30].
3.2. Studies with Confirmatory In-Vitro and/or Clinical Data
The oral Janus kinase (JAK)1/JAK2 inhibitor baricitinib, which is currently used for the treatment of rheumatoid arthritis (RA). Baricitinib was proposed by an AI algorithm due to their anti-cytokine effects and as an inhibitor of host cell viral propagation (proven anti-inflammatory effects and predicted antiviral effects) [32,33][17][18]. The aberrant activation of the JAK-STAT (signal transducer and activator of transcription) signaling pathway is related to the regulation of the immune system response. The aberrant activation of the JAK-STAT occurs in certain diseases, such as RA or psoriatic arthritis. Diverse cytokines are involved in the activation of JAK/STAT, such as pro-inflammatory cytokines (e.g., IL6, IL12, IL23 or TNF). Cytokine binding to JAK/STAT activate the signaling cascade, which leads to the activation of JAKs. This mechanism may explain the potential benefit of using JAK inhibitors, such as baricitinib in the treatment of COVI-2019, regarding cytokines may be increased (e.g., cytokine storm) in COVID-2019 [7,8,15][7][8][31].
The AI platform (BenevolentAI) predicted that baricitinib might also inhibit Numb-associated kinases (NAKs), which are important for viral entry into a cell (i.e., prediction of antiviral effects). Particularly, inhibition of NAKs led to reduced viral infectivity with baricitinib using human primary liver spheroids. In vitro pharmacology studies of baricitinib across relevant leukocyte subpopulations and in vivo pharmacokinetics data demonstrated the inhibition of signaling of cytokines implicated in COVID-19 infection. The pharmacologic effect of barictinib is supported by both in-vitro (dual antiviral and anti-inflammatory activities in liver organoids; human liver spheroids infected with SARSCoV-2) and clinical data (4 hospitalized patients).
Additionally, clinical, and radiologic recovery, a quick diminution in SARS-CoV-2 viral load, inflammatory markers reduced (e.g., C-reactive protein or interleukin-6), and IL-6 levels were achieved in the treatment of 4 hospitalized patients with COVID-2019. Compassionate use was authorized, regarding the previous approved application in RA [32,33][17][18].
References
- Dhar Chowdhury, S.; Oommen, A.M. Epidemiology of COVID-19. J. Dig. Endosc. 2020, 11, 3–7.
- WHO. Archived: WHO Timeline—COVID-19. 2020. Available online: https://www.who.int/news/item/27-04-2020-who-timeline---covid-19 (accessed on 18 July 2021).
- WHO. WHO Coronavirus (COVID-19) Dashboard. Available online: https://covid19.who.int/ (accessed on 18 July 2021).
- Yang, L.; Xie, X.; Tu, Z.; Fu, J.; Xu, D.; Zhou, Y. The signal pathways and treatment of cytokine storm in COVID-19. Signal Transduct. Target. Ther. 2021, 6, 255.
- Gurung, A.B.; Ali, M.A.; Lee, J.; Farah, M.A.; Al-Anazi, K.M. An Updated Review of Computer-Aided Drug Design and Its Application to COVID-19. BioMed Res. Int. 2021, 2021, 8853056.
- Pushpakom, S.; Iorio, F.; Eyers, P.A.; Escott, K.J.; Hopper, S.; Wells, A.; Doig, A.; Guilliams, T.; Latimer, J.; McNamee, C.; et al. Drug repurposing: Progress, challenges and recommendations. Nat. Rev. Drug Discov. 2019, 18, 41–58.
- Liu, B.M.; Martins, T.B.; Peterson, L.K.; Hill, H.R. Clinical significance of measuring serum cytokine levels as inflammatory biomarkers in adult and pediatric COVID-19 cases: A review. Cytokine 2021, 142, 155478.
- Liu, B.M.; Hill, H.R. Role of Host Immune and Inflammatory Responses in COVID-19 Cases with Underlying Primary Immunodeficiency: A Review. J. Interferon Cytokine Res. 2020, 40, 549–554.
- Aleem, A.; Akbar Samad, A.B.; Slenker, A.K. Emerging Variants of SARS-CoV-2 And Novel Therapeutics Against Coronavirus (COVID-19); StatPearls Publishing: Treasure Island, FL, USA, 2021. Available online: https://www.ncbi.nlm.nih.gov/books/NBK570580/#:~:text=Available%20from%3A%20https%3A//www.ncbi.nlm.nih.gov/books/NBK570580/ (accessed on 31 July 2021).
- Centers for Disease Control and Prevention. SARS-CoV-2 Variant Classifications and Definitions. Available online: https://www.cdc.gov/coronavirus/2019-ncov/variants/variant-info.html (accessed on 18 July 2021).
- EMA. Treatments and Vaccines for COVID-19. Available online: https://www.ema.europa.eu/en/human-regulatory/overview/public-health-threats/coronavirus-disease-covid-19/treatments-vaccines-covid-19 (accessed on 18 July 2021).
- Page, M.J.; McKenzie, J.E.; Bossuyt, P.M.; Boutron, I.; Hoffmann, T.C.; Mulrow, C.D.; Shamseer, L.; Tetzlaff, J.M.; Akl, E.A.; Brennan, S.E.; et al. The PRISMA 2020 statement: An updated guideline for reporting systematic reviews. BMJ 2021, 372, n71.
- Ke, Y.-Y.; Peng, T.-T.; Yeh, T.-K.; Huang, W.-Z.; Chang, S.-E.; Wu, S.-H.; Hung, H.-C.; Hsu, T.-A.; Lee, S.-J.; Song, J.-S.; et al. Artificial intelligence approach fighting COVID-19 with repurposing drugs. Biomed. J. 2020, 43, 355–362.
- Gysi, D.M.; Do Valle, Í.; Zitnik, M.; Ameli, A.; Gan, X.; Varol, O.; Ghiassian, S.D.; Patten, J.J.; Davey, R.A.; Loscalzo, J.; et al. Network medicine framework for identifying drug-repurposing opportunities for COVID-19. Proc. Natl. Acad. Sci. USA 2021, 118, e2025581118.
- Abdulla, A.; Wang, B.; Qian, F.; Kee, T.; Blasiak, A.; Ong, Y.H.; Hooi, L.; Parekh, F.; Soriano, R.; Olinger, G.G.; et al. Project IDentif.AI: Harnessing Artificial Intelligence to Rapidly Optimize Combination Therapy Development for Infectious Disease Intervention. Adv. Ther. 2020, 3, 2000034.
- Blasiak, A.; Lim, J.J.; Seah, S.G.K.; Kee, T.; Remus, A.; Chye, D.H.; Wong, P.S.; Hooi, L.; Truong, A.T.L.; Le, N.; et al. IDentif.AI: Rapidly optimizing combination therapy design against severe Acute Respiratory Syndrome Coronavirus 2 (SARS-Cov-2) with digital drug development. Bioeng. Transl. Med. 2020, 6, e10196.
- Schultz, M.B.; Vera, D.; Sinclair, D.A. Can artificial intelligence identify effective COVID-19 therapies? EMBO Mol. Med. 2020, 12, e12817.
- Stebbing, J.; Krishnan, V.; De Bono, S.; Ottaviani, S.; Casalini, G.; Richardson, P.J.; Monteil, V.; Lauschke, V.M.; Mirazimi, A.; Youhanna, S.; et al. Mechanism of baricitinib supports artificial intelligence-predicted testing in COVID-19 patients. EMBO Mol. Med. 2020, 12, e12697.
- Nayarisseri, A.; Khandelwal, R.; Madhavi, M.; Selvaraj, C.; Panwar, U.; Sharma, K.; Hussain, T.; Singh, S.K. Shape-based Machine Learning Models for the Potential Novel COVID-19 Protease Inhibitors Assisted by Molecular Dynamics Simulation. Curr. Top. Med. Chem. 2020, 20, 2146–2167.
- Kim, J.; Zhang, J.; Cha, Y.; Kolitz, S.; Funt, J.; Chong, R.E.; Barrett, S.; Kusko, R.; Zeskind, B.; Kaufman, H. Advanced bioinformatics rapidly identifies existing therapeutics for patients with coronavirus disease-2019 (COVID-19). J. Transl. Med. 2020, 18, 257.
- Das, G.; Das, T.; Chowdhury, N.; Chatterjee, D.; Bagchi, A.; Ghosh, Z. Repurposed drugs and nutraceuticals targeting envelope protein: A possible therapeutic strategy against COVID-19. Genomics 2021, 113 Pt 2, 1129–1140.
- Rajput, A.; Thakur, A.; Mukhopadhyay, A.; Kamboj, S.; Rastogi, A.; Gautam, S.; Jassal, H.; Kumar, M. Prediction of repurposed drugs for Coronaviruses using artificial intelligence and machine learning. Comput. Struct. Biotechnol. J. 2021, 19, 3133–3148.
- Li, Z.; Yao, Y.; Cheng, X.; Chen, Q.; Zhao, W.; Ma, S.; Li, Z.; Zhou, H.; Li, W.; Fei, T. A computational framework of host-based drug repositioning for broad-spectrum antivirals against RNA viruses. iScience 2021, 24, 102148.
- McCoy, K.; Gudapati, S.; He, L.; Horlander, E.; Kartchner, D.; Kulkarni, S.; Mehra, N.; Prakash, J.; Thenot, H.; Vanga, S.; et al. Biomedical Text Link Prediction for Drug Discovery: A Case Study with COVID-19. Pharmaceutics 2021, 13, 794.
- Chakravarty, K.; Antontsev, V.G.; Khotimchenko, M.; Gupta, N.; Jagarapu, A.; Bundey, Y.; Hou, H.; Maharao, N.; Varshney, J. Accelerated Repurposing and Drug Development of Pulmonary Hypertension Therapies for COVID-19 Treatment Using an AI-Integrated Biosimulation Platform. Molecules 2021, 26, 1912.
- Kadioglu, O.; Saeed, M.; Greten, H.J.; Efferth, T. Identification of novel compounds against three targets of SARS CoV-2 coronavirus by combined virtual screening and supervised machine learning. Comput. Biol. Med. 2021, 133, 104359.
- Artigas, L.; Coma, M.; Matos-Filipe, P.; Aguirre-Plans, J.; Farrés, J.; Valls, R.; Fernandez-Fuentes, N.; De La Haba-Rodriguez, J.; Olvera, A.; Barbera, J.; et al. In-silico drug repurposing study predicts the combination of pirfenidone and melatonin as a promising candidate therapy to reduce SARS-CoV-2 infection progression and respiratory distress caused by cytokine storm. PLoS ONE 2020, 15, e0240149.
- Wang, Z.; Li, L.; Song, M.; Yan, J.; Shi, J.; Yao, Y. Evaluating the Traditional Chinese Medicine (TCM) Officially Recommended in China for COVID-19 Using Ontology-Based Side-Effect Prediction Framework (OSPF) and Deep Learning. J. Ethnopharmacol. 2021, 272, 113957.
- Acharya, A.; Agarwal, R.; Baker, M.B.; Baudry, J.; Bhowmik, D.; Boehm, S.; Byler, K.G.; Chen, S.Y.; Coates, L.; Cooper, C.J.; et al. Supercomputer-Based Ensemble Docking Drug Discovery Pipeline with Application to Covid-19. J. Chem. Inf. Model. 2020, 60, 5832–5852.
- Kaushal, K.; Sarma, P.; Rana, S.V.; Medhi, B.; Naithani, M. Emerging role of artificial intelligence in therapeutics for COVID-19: A systematic review. J. Biomol. Struct. Dyn. 2020, 10, 1–16.
- Gatti, M.; Turrini, E.; Raschi, E.; Sestili, P.; Fimognari, C. Janus Kinase Inhibitors and Coronavirus Disease (COVID)-19: Rationale, Clinical Evidence and Safety Issues. Pharmaceuticals 2021, 14, 738.
More