Glioblastoma (GBM) is one of the most aggressive brain malignancies with high incidences of developing treatment resistance, resulting in poor prognoses. Glioma stem cell (GSC)-derived exosomes are important players that contribute to GBM tumorigenesis and aggressive properties.
- GBM-N019
- exosome
- glioma stem cell
- palbociclib
- drug resistance
- combination therapy
1. Introduction
Despite advancements in the knowledge and understanding of the mechanisms involved in the tumor biology of glioblastoma (GBM) and various treatment modalities over the last few decades, GBM remains one of the deadliest and the most common primary brain tumors, with tremendously poor prognoses[1] [1]. The success of clinical trials of new chemotherapies and standard therapies has been disappointing[2] [2] due to several factors including the drug delivery limiting features of blood–brain barrier (BBB), GBM immune-suppressive microenvironments, and structural fragility of the brain[3] [3]. Furthermore, the existence of glioma-initiating cells (GICs) a type of glioma stem cell (GSC), mediates treatment failure, tumor recurrence, and invasive phenotypes of GBM [4]. As a carrier of oncogenes/proteins and other genetic information, exosomes are involved in the conversion of non-GSCs to GSCs and participate in stabilizing the GSC phenotypic integrity[5] [5]. GBM is also characterized by complex and heterogeneous genotypes which limit the efficacy of drugs that target specific oncogenic signaling axes[6] [6]. Thus, developing a multi-oncotarget treatment modality that targets GSCs and exosomes may improve the devastating prognoses of GBM.
The phosphatidylinositol-4,5-biphosphate 3-kinase (PI3K)/Akt/mammalian target of rapamycin (mTOR) signaling pathway has emerged as one of the most deregulated oncogenic pathways that contribute to aggressive phenotypes, drug resistance, and poor prognoses in GBM patients[7] [7]. However, due to several limiting factors [8][9][10][11][8,9,10,11], efforts to target this signaling axis failed to improve the prognoses of GBM patients [8]. Importantly, the inability of mTOR inhibitors to target mTORC2 epitomizes another major clinical limitation of targeted therapy [12][12]. Therefore, targeting mTORC2 would overcome the limitations of mTORC1 inhibitors and provide a sound therapeutic strategy for GBM. This is supported by preclinical evidence of the critical roles of mTORC2 in GBM biology and the extenuating effect of targeting mTORC2 on GBM growth, invasive phenotypes, and drug resistance[13][14][15][16] [13,14,15], thus paving the way for personalized and targeted therapy.
Cyclin-dependent kinases (CDKs) are members of the serine/threonine protein kinase family that regulates cell division and transcription [16,17]. Unrestricted cell cycle progression and high cellular growth due to aberrant CDK4/6 signaling have been identified as hallmarks of astrocytic tumorigenesis and glioma progression in most GBM cases [17][18][19][18]. Hyper-expression of CDK4/6 was also documented in several other cancer types [20][21][22][23] [19,20,21,22,23]. Although mono-therapeutic inhibitors that target CDK4/6 signaling pathways have been developed[24][25][26] [24,25], their efficacy in GBM remains disappointing [18], thus accentuating the need for synergistic contributions from other agents.
Summing up the above literature with the clinical data from The Cancer Genome Atlas (TCGA) database strongly suggests that CDK6/mTOR/STAT3 overexpression is correlated with a high glioma grade, lower survival, and poor prognosis in glioblastoma patients [18][23][18,23]. The presence of GSCs and oncogene delivery features of exosomes [5], together with aberrations of CDK6/mTOR/STAT3 oncogenic pathways, concomitantly contribute to aggressive phenotypes and the failure of therapeutic strategies against GBM[25] [25]. New therapeutic strategies are urgently needed to address all the challenges mentioned above to improve patients’ survival.
Palbociclib is an oral selective inhibitor of CDK4/6, which leads to phosphorylation of RB1 and cell-cycle arrest [26]. RB1 status, therefore, becomes a determinant of tumor sensitivity to palbociclib therapy. Disappointedly, about 11% of GBM show complete loss of RB1 transcript expression [27], rendering them resistant to palbociclib [28]. Clinical studies have also demonstrated that CDK4/6 inhibitor alone showed sub-optimal efficacy for recurrent glioblastoma [29][30][31][29,30,31]. Thus, in combination with other therapies, palbociclib has been vigorously tested and proven effective in some patients [28][30][31][32][33][34][35][36][28,31,32,33,34,35,36]. Specifically, mTOR inhibitor with palbociclib showed increased efficacy against GBM [37][38][37,38]. In addition, there are several ongoing trials testing combinations of palbociclib with immunotherapy, including avelumab and pembrolizumab (NCT02778685; NCT02779751; and NCT03147287) [39]. Collectively, these findings strongly suggested that targeting mTOR/CDK6 associated signaling is a potential new target for developing GBM therapeutics.
Anthraquinone-derived heterocyclic scaffolds have been explored for drug design, discovery, and development[40] [40], and drug candidates from this class have demonstrated antitumor activities in various studies [41][41,42,43,44,45]. GBM-N019 is a novel member of a series of anthraquinone-derived, tetraheterocylic azathioxanthone derivatives. Herein, we demonstrated for the first time through a series of in vitro and in vivo studies that GBM-N019 significantly compromised the viability and tumorigenic features of GBM cells via downregulation of STAT3, Akt, mTOR, nuclear factor (NF)-κB, and CDK6 signaling networks. We also found that GBM-N019 halted the exosomal cargo delivery of Akt, mTOR, p-mTOR, and RAB27A, and attenuated the tumorsphere-derived exosomes (exosphere; Exosp) mediated drug resistance and aggressive phenotypes of GBM.
2. Results
2. mTOR, STAT3, and CDK6 Are Key Oncogenic Signatures of Disease Progression, Therapy Failure, and Poor Prognosis in GBM Patients
2.1. mTOR, STAT3, and CDK6 Are Key Oncogenic Signatures of Disease Progression, Therapy Failure, and Poor Prognosis in GBM Patients
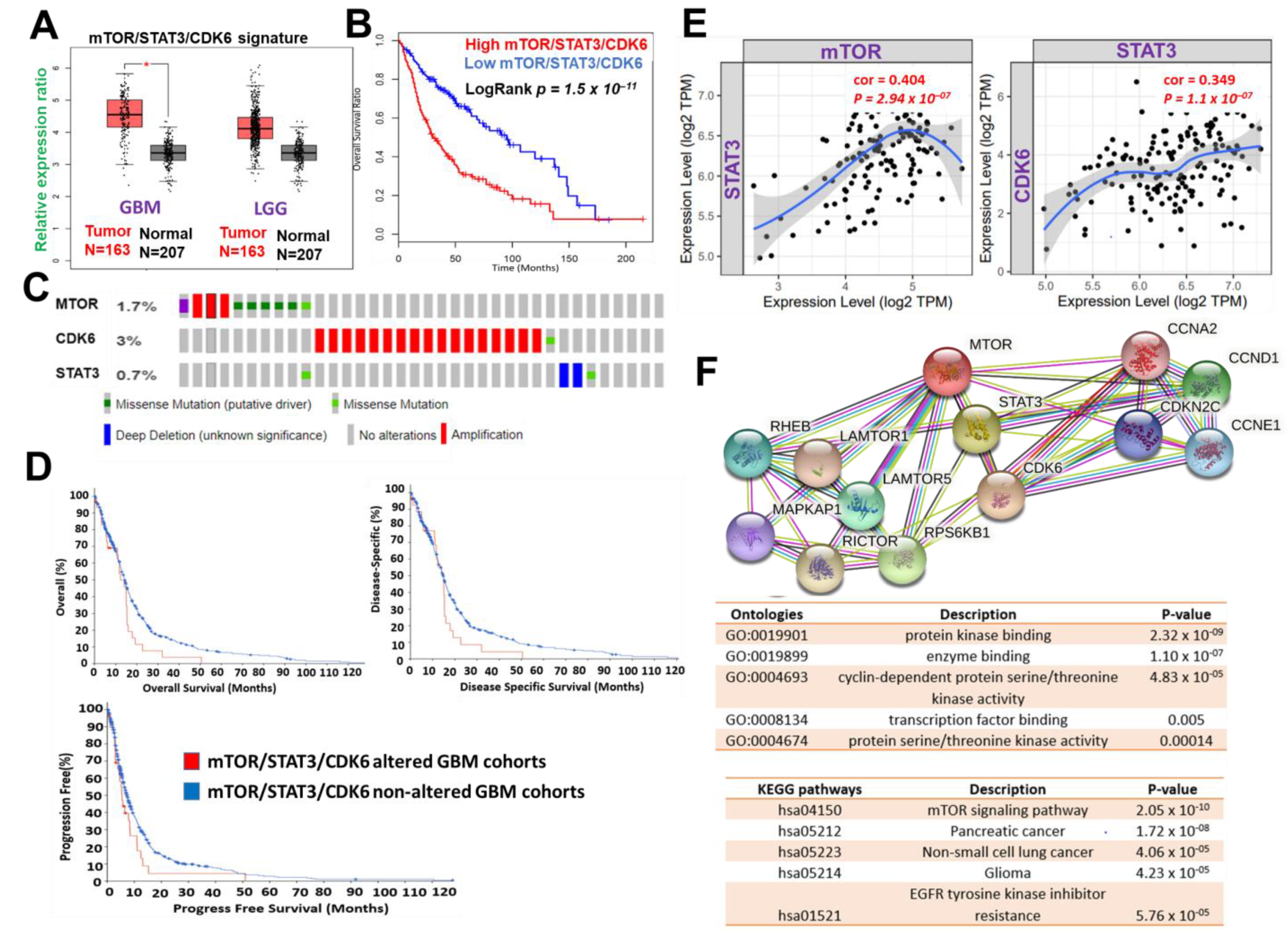 Figure 1. mTOR, STAT3 and CDK6 is an important oncogenic signature of disease progression, therapy failure, and poor prognosis in GBM patients. (A) Cumulative mTOR, STAT3, and CDK6 expression levels in the patients with GBM and low-grade glioma (LGG) from TCGA and GTEx datasets. (B) Kaplan–Meier plots of the cumulative survival of GBM patients. (C) Frequency of genetic alterations of mTOR, STAT3 and CDK6 signature in glioblastoma (TCGA, PanCancer Atlas) dataset. (D) Kaplan–Meier plots of the overall survival, disease-specific, and disease progressive free survival of GBM cohorts with genetically altered mTOR, STAT3 and CDK6 signature. (E) Correlation analysis of the expression between STAT3 and mTOR (left) and between CDK6 and STAT3 (right) from GBM TCGA databases. Both p-values and Spearman’s rank correlation coefficient (cor) are indicated. (F) Protein–protein interaction networks of mTOR, STAT3 and CDK6 signature (upper panel) and the enriched KEGG pathways and gene ontologies in the mTOR, STAT3 and CDK6 clustering network.
Figure 1. mTOR, STAT3 and CDK6 is an important oncogenic signature of disease progression, therapy failure, and poor prognosis in GBM patients. (A) Cumulative mTOR, STAT3, and CDK6 expression levels in the patients with GBM and low-grade glioma (LGG) from TCGA and GTEx datasets. (B) Kaplan–Meier plots of the cumulative survival of GBM patients. (C) Frequency of genetic alterations of mTOR, STAT3 and CDK6 signature in glioblastoma (TCGA, PanCancer Atlas) dataset. (D) Kaplan–Meier plots of the overall survival, disease-specific, and disease progressive free survival of GBM cohorts with genetically altered mTOR, STAT3 and CDK6 signature. (E) Correlation analysis of the expression between STAT3 and mTOR (left) and between CDK6 and STAT3 (right) from GBM TCGA databases. Both p-values and Spearman’s rank correlation coefficient (cor) are indicated. (F) Protein–protein interaction networks of mTOR, STAT3 and CDK6 signature (upper panel) and the enriched KEGG pathways and gene ontologies in the mTOR, STAT3 and CDK6 clustering network.3. mTOR, STAT3, and CDK6 Are Druggable Targets for a Novel Drug-like Multitarget Small Molecule (GBM-N019)
.2. mTOR, STAT3, and CDK6 Are Druggable Targets for a Novel Drug-like Multitarget Small Molecule (GBM-N019)
Computational study revealed that the GBM-N019 is BBB permeant (Figure 2A), it has acceptable absorption, distribution, metabolism, and excretion (ADME) characteristics and met all criteria of Lipinski’s rule of five. Furthermore, with computer-based drug target predictions and using GBM-N019 as a query molecule, we identified a number of targetable proteins, most of which were previously implicated in the poor prognosis of GBM. Among these predicted target proteins, enzymes (33.3%), kinases (26.7%), proteases (13.3%), electrochemical transporters (13.3%), and family A G protein-coupled receptors (13.3%) were the most repartitioned targeted classes . Specifically, three members of cyclin-dependent kinases (CDK4, -6, and -9) and three members of the PI3K family (PI3K p110-delta subunit (PIK3CD), PI3K p110-beta subunit (PIK3CB), and PI3K p110-gamma subunit (PIK3CG)) were predicted to be targeted by GBM-N019. Additionally, using GBM-N019 as the query molecules, mTOR, NF-κB, and STAT6 appeared as part of the topmost targets . Molecular docking studies revealed that GBM-N019 docked well into the binding cavity of (Figure 2B), mTOR (Figure 2C), and STAT3 (Figure 2D) with binding affinities of −7.6, −8.1, and −6.9 Kcal/mol, respectively. The ligand receptors complex was bound by several hydrogen bonding, alkyl interactions. Furthermore, the complex was stabilized by various hydrophobic contacts and several Van der Waal forces surrounding the ligand backbone (Figure 1B). Interestingly, GBM-N019 bind to CDK6 with the same affinity that palbociclib has for CDK6 (ΔG = −8.1 Kcal/mol); however, the affinities of GBM-N019 for mTOR and STAT3 are lower than that of their respective standard inhibitors; Dactolisib (ΔG = −9.2 Kcal/mol vs. −7.6 Kcal/mol) and SH-4-54 (ΔG = −7.3 Kcal/mol vs. −6.9 Kcal/mol) . Collectively, our computational study revealed that GBM-N019 is a novel drug candidate as an inhibitor of mTOR/STAT3/CDK6 signature.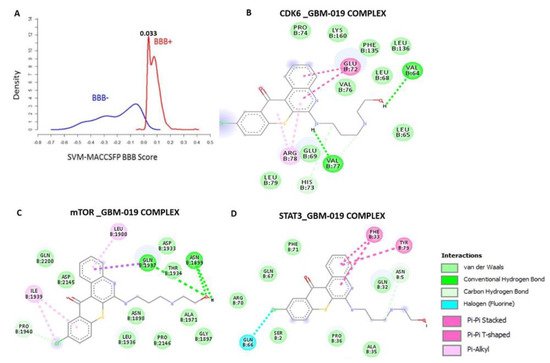
4. GBM-N019 Curbed the Viability and Tumorigenic Features of GBM Cells via Downregulation of NF-κB/Akt/mTOR, STAT3, and CDK6 Signaling In Vitro
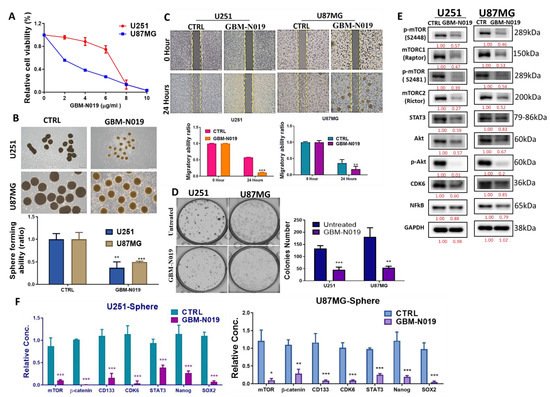
3.3. GBM-N019 Curbed the Viability and Tumorigenic Features of GBM Cells via Downregulation of NF-κB/Akt/mTOR, STAT3, and CDK6 Signaling In Vitro
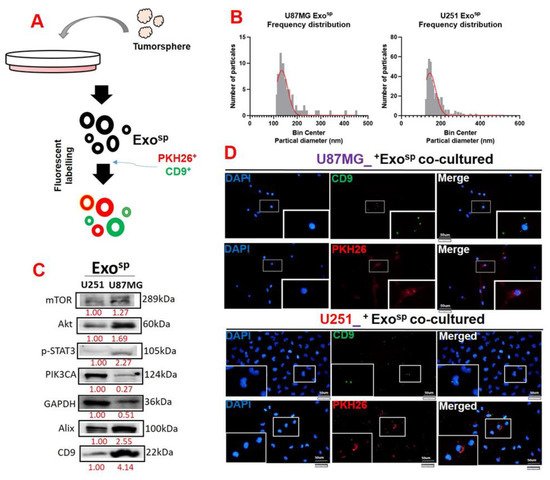
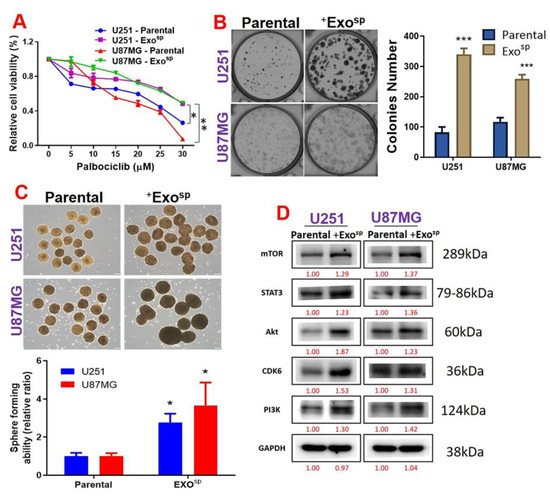
5. Tumorsphere-Derived Exosomal Cargo of Oncogenes Mediated Treatment Resistance and Aggressive Phenotypes of GBM
3.4. Tumorsphere-Derived Exosomal Cargo of Oncogenes Mediated Treatment Resistance and Aggressive Phenotypes of GBM