Your browser does not fully support modern features. Please upgrade for a smoother experience.
Please note this is a comparison between Version 1 by Antonio Pineda-Lucena and Version 2 by Camila Xu.
Breast cancer (BC) is the most frequently diagnosed tumor and the leading cause of cancer deaths in women worldwide.。
- metabolism
- breast cancer
- multi-omics
- biomarkers
- early diagnosis
1. Introduction
Breast cancer (BC) is the most frequently diagnosed tumor and the leading cause of cancer deaths in women worldwide [1]. The year 2020 saw an estimated 2.3 million new cases of BC (11.7% of all cancer cases), with 685,000 deaths worldwide [1]; however, advances in population screening and early treatment (among other factors) have supported a steady decrease in BC mortality [2][3][2,3]. Unfortunately, figures from the American Cancer Society place the five-year survival rate after diagnosis of metastatic BC at 27%, a low value considering the 99% five-year survival rate for localized disease [4]. Therefore, early BC detection represents a crucial step in reducing disease mortality [5]. BC screening currently relies on mammography, a non-invasive strategy primarily performed in women between 50 to 69 years of age that has prompted a reduction in BC mortality [6]. Nevertheless, this approach suffers from several limitations, including false-positive reporting and overdiagnosis [6][7][8][9][10][6,7,8,9,10]. Ultrasound, magnetic resonance imaging, and computed tomography can overcome such problems thanks to their high sensitivity; however, the elevated costs associated with these tools make this approach less accessible. Thus, we still lack alternative methods for the accurate, non-invasive, and low-cost diagnosis of early-stage BC.
BC is a highly heterogeneous disease from a molecular perspective and is primarily characterized by the overexpression of the HER2 growth factor, estrogen receptor (ER), and progesterone receptor (PR) and mutations in the BRCA1/2 genes, with the latter associated with a higher risk of developing BC [11]. Former classifications of BC tumors employed tumor size, histological grade, immunohistochemistry of ER/PR status, and the amplification of HER2. The addition of gene expression profiling to these molecular features has resulted in the classification currently used by the European Society for Medical Oncology as a clinical guideline for BC diagnosis, follow-up, and treatment [12][13][12,13]. This system classifies BC tumors into four major intrinsic molecular subgroups: luminal A (ER+ and/or PR+, HER2-, low Ki67), luminal B (ER+ and/or PR+, HER2+ or HER2- with high Ki67), basal-like (ER/PR-, HER2-), and HER2-enriched (ER/PR-, HER2+). Luminal A tumors (the low-grade group) are the most common BC subtype, comprising over 60–70% of all cases. Meanwhile, basal-like tumors, with an ~80% overlap with highly proliferative triple-negative breast cancer (TNBC) [14], exhibit aggressive behavior and suffer from poor prognosis [15]. Although ER+ tumors present lower recurrence rates within the first five years, over 50% of tumor recurrences occur after this time and cause most BC-related deaths [16][17][16,17]. Each BC subtype has a characteristic biological profile, prognosis, and treatment strategy [18][19][20][21][22][18,19,20,21,22], and several scoring systems aid prognosis and treatment decision-making processes. Unfortunately, systems based on different molecular features related to tumor biology, including histological type, grade, lymphovascular invasion, and marker status, do not accurately reflect BC subtype heterogeneity or specific patient subtypes [23]. Thus, enormous efforts have been devoted to classifying heterogeneous BC subtypes into molecular subtypes that guide treatment decisions [24][25][26][27][28][24,25,26,27,28].
2. Multi-Omics Studies of BC Prognosis
Metabolic deregulation can impact various molecular processes (e.g., cell proliferation, apoptosis, migration, and invasion) that contribute to tumor progression [29][30][31][32][44,108,109,110] and influence cancer patient survival (Figure 2) [33][111].

Figure 2. Schematic representation of the impact of metabolic changes on essential molecular processes associated with tumor progression and patient survival. Created with BioRender.com.
Several studies have reported associations between metabolic alterations and BC patient survival based on single omic analysis, including genomic [34][112], transcriptomic [35][113], proteomic [36][46], and metabolomic [37][38][39][56,114,115] studies in tissue [36][34][35][46,112,113] and serum [37][38][39][56,114,115] samples. However, additional studies based on the integration of data from multi-omics analyses have provided more accurate information regarding the molecules involved in metabolic rewiring associated with BC progression. Of the seven studies following a multi-omics approach to identify metabolic alterations associated with BC prognosis (Table 3), most relied on the analysis of tissue sample analysis and the integration of transcriptomic and metabolomic datasets. Overall, BC patient survival associated with the altered expression of enzymes involved in nucleotide, lipid, and amino acid metabolism.
Previous studies have demonstrated that nucleotide biosynthesis plays a vital role in BC [47][48][49][50][51][79,80,83,84,117], and could represent a promising therapeutic strategy [52][53][54][54,85,118]. Notably, two of the multi-omics studies included in Table 3 established an inverse correlation between the expression levels of genes involved in de novo purine and pyrimidine syntheses and BC patient survival [41][40][61,65]. Putluri et al. performed an in silico analysis to evaluate the association between omics-based enrichment and patient survival, using ten independent gene expression data sets to select clinically relevant prognostic biomarkers [40][65]. Kaplan–Meyer curves revealed an association between increased expression of pyrimidine metabolism-related genes and shorter metastasis-free survival across all BC and within the subset of ER + tumors. RRM2, a critical gene in pyrimidine metabolism displaying elevated expression in aggressive BC [55][119], has prognostic relevance in BC [56][57][120,121] when combined with proliferation markers [58][122]. RRM2 expression distinguished good vs. poor survival within the entire BC patient group in this multi-omics study, including a significant proportion of luminal A subtype typically considered to have better survival outcomes [48][80]. Luo and colleagues integrated metabolomic and transcriptomic analysis to confirm the association between alterations in nucleotide metabolism and BC patient survival in a TCGA cohort of patients [41][61]. The authors observed a significant correlation between poor survival of BC patients and changes to the expression of RRM2 and adenosine monophosphate deaminase 1 (AMPD1), a key enzyme in de novo purine synthesis. These enzymes have been postulated as promising therapeutic targets in different tumor types, including BC [40][53][59][60][61][62][65,85,123,124,125,126].
Table 3.
Multi-omics studies focused on identifying metabolic alterations associated with BC prognosis.
| Study | Sample | Omics Data | Major Findings * | |||||
|---|---|---|---|---|---|---|---|---|
| Putluri et al. [40] | Putluri et al. [65] | Cell lines + Tissue | M+T | ↑ | RRM2 | (pyrimidine metabolism) | ||
| Luo et al. [41] | Luo et al. [61] | Blood + Tissue | M+T | ↑ | RRM2 | (pyrimidine metabolism) and ↓ | AMPD1 | (de novo purine metabolism) |
| Iqbal et al. [42] | Iqbal et al. [59] | Tissue | M+T | ↑ | CBX2 | and ↓ | CBX7 | (glycolysis) |
| Camarda et al. [43] | Camarda et al. [102] | Cell lines + Tissue | M+T | ↓ ACC2 (FAO) | ||||
| Kang et al. [44] | Kang et al. [116] | Cell lines | L+T | ↓ | ELOVL2 | (lipid synthesis) | ||
| Terunuma et al. [45] | Terunuma et al. [107] | Tissue | M+T+P+E | ↑ 2HG, SAM and SAH ↑ | IDH2 | (glutamine metabolism) | ||
| Budczies et al. [46] | Budczies et al. [106] | Tissue | M+T | ↓ | ABAT | , ↑ β-alanine (β-alanine metabolism) |
ABAT: 4-aminobutyrate aminotransferase, ACC2: acetyl-CoA carboxylase 2, AMPD1: adenosine monophosphate deaminase 1, CBX2: chromobox 2, CBX7: chromobox 7, E: epigenomics, ELOVL2: ELOVL fatty acid elongase 2, FAO: fatty acid oxidation, L: lipidomics, IDH2: isocitrate dehydrogenase (NADP(+)) 2, M: metabolomics, P: proteomics, RRM2: ribonucleotide reductase regulatory subunit M2, SAH: S-adenosylhomocysteine, SAM: S-adenosyl- methionine, T: transcriptomics, 2HG: 2-hydroxyglutarate. * Direction of metabolic alterations directly correlated with worse BC patients’ outcomes.
Iqbal et al. established antagonistic roles of CBX2 and CBX7 in metabolic reprogramming of BC and an association with BC patient survival [42][59]. The authors described a significant correlation between higher CBX2 and lower CBX7 mRNA levels and worse BC prognosis, which agrees with previous findings that correlated the CBX2 or CBX7 expression with overall patient survival [63][64][65][66][127,128,129,130].
As for the alterations in lipid metabolism, Camarda and colleagues followed a targeted metabolomics approach and reported the dramatic upregulation of FAO intermediates in a MYC-driven model of TNBC [43][102]. To characterize a potential association between FAO gene expression and prognosis in TNBC, the authors performed a univariate analysis of 336 fatty acid metabolism genes on a patient cohort with long-term distant recurrence-free survival data. The analysis revealed that decreased ACACB (acetyl-CoA carboxylase 2, ACC2) expression levels associated with worse prognoses in all BC and TNBC patients. Subsequent studies also described significant associations between increased levels of ACC2 and better BC prognosis [67][68][69][70][131,132,133,134]. Kang et al. conducted a multi-layered lipidomics and transcriptomics analysis to describe the rewiring of the BC lipidome during malignant transformation [44][116]. Analyses in a spheroid-induced epithelial-mesenchymal transition (EMT) model demonstrated a dramatic reduction in the ratio of C22:6n3 (docosahexaenoic acid, DHA) to C22:5n3 in spheroid cells, similarly to the down-regulation of ELOVL2, a process associated with the induction of metastatic characteristics in BC cells. The authors examined the relationship between ELOVL2 expression and metastatic relapse-free in a BC cohort with a follow-up of ten years, resulting in the discovery of an association between lower ELOVL2 expression levels and shorter metastasis-free survival and higher tumor grade [44][116]. A recent study investigating the molecular mechanisms of tamoxifen resistance in BC confirmed these findings and described lower ELOVL2 expression in tamoxifen-resistant models and ELOVL2 downregulation in patients with tamoxifen resistance [71][135].
Additional studies have revealed a correlation between alterations in the levels of genes and metabolites involved in amino acid metabolism and BC prognosis. Terunuma and coworkers identified a subset of BC tumors accumulating high levels of 2-hydroxyglutarate (2HG). Further analyses revealed the presence of a subgroup of BC patients with significantly decreased survival characterized by an exceptionally high accumulation of 2HG, reduced DNA methylation at the isocitrate dehydrogenase (IDH2) locus, increased IDH2 expression, and increased levels of S-adenosyl- methionine (SAM) and S-adenosylhomocysteine (SAH) [45][107]. Previous studies reported an accumulation of the oncometabolite 2HG in different tumor types, including BC [72][67], glioma [73][136], and leukemia [74][137]. In another study, concerning amino acid metabolism, Budczies et al. described an association between alterations in the metabolism of β-alanine and shorter recurrence-free survival of BC patients [46][106]. Specifically, the authors demonstrated that lower expression levels of 4-aminobutyrate aminotransferase (ABAT), which negatively correlated with the concentration of β-alanine, indicated worse prognoses in BC patients. A similar study reported decreased ABAT expression in more aggressive BC subtypes, which correlated with an increased risk of metastasis and shorter overall, relapse-free, and distant metastasis-free survival [75][138]. Finally, Jansen et al. correlated ABAT downregulation with poor progression-free and metastasis-free survival in tamoxifen-treated patients [76][139].
34. Multi-Omics Studies and Novel BC Treatment Strategies
Omics-based technologies have also been used to identify novel therapeutic targets and monitor biological alterations related to BC metabolism following treatment [77][78][79][80][81][82][83][84][85][32,45,140,141,142,143,144,145,146]. The majority of studies relied on the application of metabolomics-based approaches in BC tissue [80][82][141,143] and serum [79][81][83][140,142,144] samples, although groups have evaluated transcriptomic [86][78][33,45] and proteomic [85][146] profiles in tissue samples. Various studies have described how the combination of omics approaches could characterize specific targets and foster the development of novel therapeutic strategies for specific subgroups of BC patients [87][88][89][147,148,149]. In particular, multi-omics studies have focused on identifying and validating metabolic enzymes as promising therapeutic strategies for the treatment of different BC tumors (Table 4). Figure 3 illustrates those metabolic-related genes proposed as potential therapeutic targets for treating BC patients in these studies. Overall, these findings suggest the therapeutic potential of inhibiting specific metabolic enzymes associated with glycolysis or involved in nucleotide, amino acid, and lipid metabolism in BC patients.
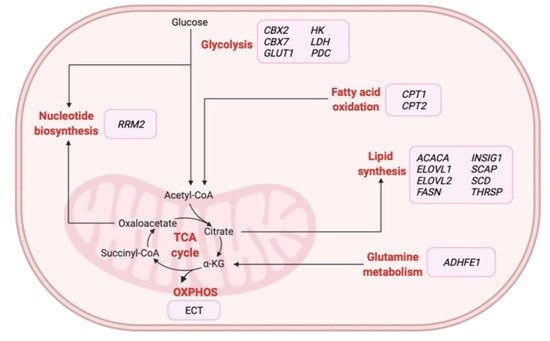

Figure 3. Overview of metabolic-related therapeutic targets for the treatment of BC patients identified from multi-omics-based studies. α-KG: alpha-ketoglutarate, ACACA: acetyl-CoA carboxylase alpha, ADHFE1: alcohol dehydrogenase iron containing 1, CBX2: chromobox 2, CBX7: chromobox 7, CPT1: carnitine palmitoyltransferase 1A, CPT2: carnitine palmitoyltransferase 2, ECT: electron transport chain, ELOVL1: ELOVL fatty acid elongase 1, ELOVL2: ELOVL fatty acid elongase 2, FASN: fatty acid synthase, GLUT1: glucose transporter 1, HK: hexokinase, INSIG1: insulin-induced gene 1, LDH: lactate dehydrogenase, OXPHOS: oxidative phosphorylation, PDC: pyruvate dehydrogenase kinase, RRM2: ribonucleotide reductase regulatory subunit M2, SCAP: SREBF chaperone, SCD: stearoyl-CoA desaturase, TCA: tricarboxylic acid, THRSP: thyroid hormone-responsive. Created with BioRender.com.
Table 4.
Multi-omics studies focused on developing new therapeutic strategies for the treatment of BC.
| Study | Omics Data | BC Subtype | Potential Targets |
|---|
| Iqbal et al. [42] | Iqbal et al. [59] | M+T | TNBC and luminal-like | CBX2 | and | CBX7 | ||||
| Gong et al. [90] | Gong et al. [69] | M+T | TNBC | FASN | and | LDH | ||||
| Mahendralingam et al. [91] | Mahendralingam et al. [66] | P+T | Basal- and luminal-like | GLUT1 | , | HK | , | LDH, | and | PDC |
| Putluri et al. [40] | Putluri et al. [65] | M+T | Basal- and luminal-like | RRM2 | ||||||
| Terunuma et al. [45] | Terunuma et al. [107] | M+T+P+E | TNBC and basal-like | ADHFE1 | ||||||
| Hilvo et al. [92] | Hilvo et al. [60] | L+T | TNBC, luminal- and basal-like |
ACACA, ELOVL1, FASN, INSIG1, SCAP, SCD | and | THRSP | ||||
| Kang et al. [44] | Kang et al. [116] | L+T | Luminal-like | ELOVL2 | ||||||
| Camarda et al. [43] | Camarda et al. [102] | M+T | TNBC and HER2 + | CPT1 | and | CPT2 |
ACACA: acetyl-CoA carboxylase alpha, ADHFE1: alcohol dehydrogenase iron containing 1, CBX2: chromobox 2, CBX7: chromobox 7, CPT1: carnitine palmitoyltransferase 1A, CPT2: carnitine palmitoyltransferase 2, E: epigenomics, ELOVL1: ELOVL fatty acid elongase 1, ELOVL2: ELOVL fatty acid elongase 2, FASN: fatty acid synthase, GLUT1: glucose transporter 1, HK: hexokinase, IDH2: isocitrate dehydrogenase (NADP(+)) 2, INSIG1: insulin-induced gene 1, L: lipidomics, LDH: lactate dehydrogenase, M: metabolomics, P: proteomics, PDC: pyruvate dehydrogenase kinase, RRM2: ribonucleotide reductase regulatory subunit M2, SCAP: SREBF chaperone, SCD: stearoyl-CoA desaturase, T: transcriptomics, TNBC: triple negative breast cancer, THRSP: thyroid hormone-responsive.
Iqbal and coworkers demonstrated that silencing CBX2 and CBX7 exhibited inverse effects on glycolysis, ATP production, viability, and proliferation [59]. CBX7 overexpression provided comparable results to CBX2 knockdown, which included decreased biomass production and reduced cell viability and proliferation. These in vitro results agreed with the findings of the transcripto-metabolomic analyses performed on BC patients and validated the roles of CBX2 and CBX7 in metabolic reprogramming of BC, highlighting the potential of these targets for the development of therapeutic strategies in BC. Of note, additional studies have provided similar results for CBX2 and CBX7 in BC [66][93][94][130,150,151], pancreatic adenocarcinoma [64][128] and metastatic prostate cancer [95][152]. Gong and coworkers evaluated the sensitivity of different BC metabolic phenotypes to metabolic inhibitors targeting glycolysis or de novo fatty acid synthesis [90][69]. The glycolytic BC phenotype displayed greater sensitivity to glycolytic inhibitors (oxamate, lactate dehydrogenase (LDH) inhibitors, and 2-deoxy-D-glucose), while inhibitors of lipid synthesis (cerulin and FASN inhibitor) exhibited higher efficacy against the lipogenic phenotype. Significantly, in vivo LDH inhibition enhanced tumor response to anti-PD-1 immunotherapy in the BC glycolytic phenotype. Previous studies have shown that LDH inhibition can suppress glycolysis [96][97][153,154] and cell proliferation [98][155] in BC cell lines. Differences in sensitivity to pharmacological inhibitors targeting glycolysis or electron transport chain (ECT) subunits were evaluated in different metabolic MECs phenotypes by Mahendralingam and colleagues [91][66]. In this study, the glycolytic phenotype displayed greater sensitivity to inhibitors targeting glucose transporter 1 (GLUT1), hexokinase (HK), LDH, and pyruvate dehydrogenase kinase (PDC), which agrees with results reported by Gong et al. [90][69]. Furthermore, studies have demonstrated that HK inhibition prevents BC growth [99][100][156,157].
The metabolic enzyme RRM2 has also been proposed as a potential therapeutic target for the treatment of BC. Putluri et al. observed that inhibiting RRM2 in BC cells significantly decreased proliferation and the expression of cell cycle genes and sensitized cells to tamoxifen treatment [40][65]. In agreement with the potential relevance of RRM2, additional studies have reported a reduction in proliferation [53][62][85,126] and tamoxifen resistance [101][158] in BC cell lines following RRM2 inhibition. Furthermore, associations between RRM2 overexpression and deterioration in BC survival have been widely reported, strongly suggesting a role as a targeted therapy for BC [41][56][57][61,120,121].
A multi-omics-based study by Terunuma et al. identified alterations associated with glutamine metabolism in a subset of BC tumors [45][107]. The authors described a subset of BC tumors with high 2HG levels and a distinct DNA methylation pattern associated with worse prognoses. Overall, studies have underscored the critical role of epigenetic-metabolomic interplay in promoting tumorigenesis [102][159]; in particular, high 2-HG levels induce epigenetic reprogramming associated with progression in different tumors [103][104][160,161]. Interestingly, the silencing of IDH2 and alcohol dehydrogenase iron containing 1 (ADHFE1), two enzymes implicated in the mitochondria-associated α-ketoglutarate–dependent production of 2HG [74][105][137,162], prompted a marked reduction of endogenous 2HG in BC cells. Furthermore, ADHFE1 loss resulted in a moderate but significant inhibition of cell cycle kinetics and reduced migration and invasion, suggesting an oncogenic role for ADHFE1 in BC. In agreement, several studies have associated high ADHFE1 expression levels with increased synthesis of 2HG and worse patient prognosis in BC [106][107][163,164].
In the context of a multi-omics study focused on lipid metabolism, Hilvo et al. conducted gene silencing experiments on seven enzymes involved in phospholipid remodeling and de novo lipid synthesis [92][60]. The results established that the individual inhibition of multiple lipid metabolism-regulating genes reduced the growth and viability of BC cell lines, which agrees with studies reporting reduced cell migration, invasion, and tumor proliferation in BC [108][109][110][165,166,167] and other tumor types [111][112][113][168,169,170] following the inhibition of specific lipid metabolism-related enzymes. Interestingly, a more recent study described FASN, another enzyme involved in de novo lipid metabolism, as a promising therapeutic target for BC treatment [114][78].
The results of a multi-omics-based study conducted by Kang et al. revealed lipid composition alterations during the EMT in BC [44][116]. The inhibition of ELOVL2 increased malignant potential, higher migration rate, and elevated colony formation. Mechanistically, downregulation of ELOVL2 increased sterol regulatory element-binding transcription factor 1 (SREBP1) expression in BC cells and activated lipogenesis, a process associated with the promotion of malignant BC phenotypes. SREBP1 is a crucial regulator of fatty acid metabolism and plays a pivotal role in the transcriptional regulation of different lipogenic genes mediating lipid synthesis [115][116][171,172]. SREBP1 overexpression has been observed in different tumor types, including BC [92][117][118][60,173,174], and supports the malignant BC phenotype [110][167]. Finally, Camarda et al. suggested the inhibition of FAO as a novel therapeutic target in a MYC-driven model of TNBC [43][102]. Based on the characterization of the effects of small-molecule inhibition and knock-down of carnitine palmitoyltransferase 1A (CPT1) and carnitine palmitoyltransferase 2 (CPT2) in BC cell lines, the authors demonstrated that FAO plays an essential role in this BC model. Furthermore, in vivo experiments demonstrated that treatment with etomoxir, a CPT1 inhibitor, significantly attenuated tumor growth in various BC models. Additional studies have also demonstrated that individual knockdown of both CPT enzymes reduces FAO metabolism and cell proliferation in different tumors [119][120][121][175,176,177]. In addition, more recent studies suggested CPT1 as a potential BC tumor target [122][123][103,178].
