Histones are alkaline proteins that package DNA into nucleosomes. H3K4me3 is highly enriched in gene promoter regions. A gain in H3K4me3 enrichment is associated with active gene transcription, open chromatin, and loss of DNA methylation. H3K4me3 has been adopted as a marker to identify transcriptionally active genes.
- assisted reproductive technologies
- ICSI
- IVF
- H3K4me3
- placenta
- umbilical cord blood
- imprint
- oxygen
- Polr2A
- KDM5A
[1]
1. Introduction
Assisted reproductive technologies (ARTs) occur at a critical window of embryo development, overlapping with extensive epigenetic reprogramming [2]. Epigenetic reprogramming plays a vital role in embryo development with the possibility of being influenced by environmental exposure [3]. Studies in mice revealed that ARTs altered the cardiovascular phenotype via epigenetic alterations under sub-optimal culturing conditions[4][5][6]. A retrospective study based on a congenital mal-formations registry in French revealed that ARTs conceived offspring have a 3-times higher risk of rare imprinting disorders associated with epigenetic dysregulation than naturally-conceived-children [6]. Epigenetic syndromes such as Beckwith-Wiedemann syndrome (BWS), Silver-Russell syndrome (SRS), Prader-Willi syndrome (PWS), and Angelman syndrome (AS) have been observed in ART-offspring[7] . A meta-analysis of 18 studies concluded that children conceived through in vitro fertilization (IVF) and intracytoplasmic sperm injection (ICSI) have an increasing risk of imprinting disorders [8]. Placentas from ART children also presented altered DNA methylation compared with those from natural conception[9] . Interestingly, ART boy has been revealed to be more susceptible to ART-treatment-associated global DNA dys-methylation[9] . However, the factors contributing to the differences observed in ART children are complex. The infertility background of ART-parents[10] , in vitro manipulation of embryo [11] and the gonadotropin stimulation[12] are possible causes for epigenetic alterations.
As important epigenetic components, histone modifications play crucial roles in responses to developmental and environmental changes [13]. Tri-methylated histone H3 lysine-4 (H3K4me3) is a common histone H3 methylated form and a marker of active transcription, which is associated with open chromatin and stable up-regulation of the gene expression downstream of H3K4me3 promoter regions [14]. It is highly enriched in promoter regions [15], involved in mammalian embryo development[16] . Acetylated histone H3 lysine-9 (H3K9ac) and acetylated histone H3 lysine-27 (H3K27ac) are common histone H3 acetylated forms associated with open chromatin and active gene transcription. They are enriched in active gene-regulatory regions, such as promoters and enhancers [17][18][19], involved in prenatal intrauterine programming [17][18]. The placenta is a multifunctional organ essential for fetal development and survival[20]. It remains unclear whether global levels of these histone modifications show differences in placentas between natural conception and ARTs.
Oxygen tension is a primary environmental factor influencing epigenetic modifications of the in vitro embryo [21]. ART laboratories have been culturing pre-implantation gametes/embryos under atmospheric O2 tension (20%), low O2 tension (5%, similar to physiologic O2 tensions in human fallopian tubes and uterus), and ultra-low O2 tension (close or less than 5%) [22]. Placentas derived from in vitro 20%-oxygen-culture condition showed significant difference in LINE1 methylation than those from in vivo conceptions, while placentas from in vitro 5%-oxygen-culture condition did not show significant differences [9]. A Cochrane review meta-analysis concluded that, compared with atmospheric O2 tension, embryos cultured in low O2 tension (5%) were better developed, bringing about higher probabilities of IVF/ICSI success, ongoing clinical pregnancy, live birth, and the birth of healthier offspring[23] . Nevertheless, the mechanisms by which low-oxygen culture improve the development of in vitro embryo are not fully clarified. It has been revealed that high-oxygen culture perturbed gene expression, metabolism, and morphology in embryos [22]. However, further studies are needed to provide detailed data on the effects of low-oxygen culture during ARTs. In addition, despite the improvement in the O2 tension of gametes/embryos culture systems, it is inevitable that ART practices are accompanied by the fluctuations of O2 tensions [24]. The impact of fluctuating O2 tensions on histone modifications has been seldom studied.
Given the available evidence, we propose two hypotheses. The first hypothesis is that histone modifications might show up as differences in comparisons between natural conception and ARTs, also in comparisons between ART-boys and ART-girls. The second is that varying oxygen conditions might influence histone modifications. We aim to test these two hypotheses and try to explore the potential regulators of histone modification.
2. Global Levels of H3K4me3 Are Reduced in Placentas from Intracytoplasmic Sperm Injection (ICSI) Than Those from Natural Conception
Firstly, we performed immunohistochemistry (IHC) staining and immune-reactive score (IRS) evaluation for H3K4me3, H3K9ac, and H3K27ac enrichment in placental tissues from natural conception, IVF, and ICSI groups. H3K4me3, H3K9ac, and H3K27ac were immunolocalized to the syncytium, fetal endothelium, and decidua. Global H3K4me3 was significantly reduced in placental syncytium (P = 0.020) and fetal endothelium (P = 0.018) from ICSI group than those from natural conception group (Figure 1). H3K4me3 showed no significant difference between natural conception and IVF groups (P > 0.05, Figure 1). H3K9ac or H3K27ac levels showed no significant difference between groups (P > 0.05).
Similar findings were recently reported by Chen and colleagues [25], who found that H3K4me3 but not H3K4me1, H3K27me3 or H3K27ac, showed differences in newborn cord blood mononuclear cell (CBMC) from natural conception and ART groups. Herein, we found ICSI placentas showed lower levels of H3K4me3 than naturally-conceived-placentas. While for IVF placentas, H3K4me3, H3K9ac or H3K27ac showed no significant difference in comparisons to the control group. Considering that IVF is a process of natural selection whereas ICSI manipulation is invasive, there is biologic plausibility to support our findings. Notably, H3K4me3 is a promising biomarker that could convey important information. A gain in H3K4me3 enrichment is associated with loss of DNA methylation, open chromatin, and active gene transcription [14][26]; thus, it has been adopted as a marker to identify genes that were transcriptionally active [27]. Besides, H3K4me3 is the most affected histone modification in the in vitro culture system [28]; a previous study suggested that H3K4me3 may potentially serve as a marker for evaluating the influence of ARTs [25].
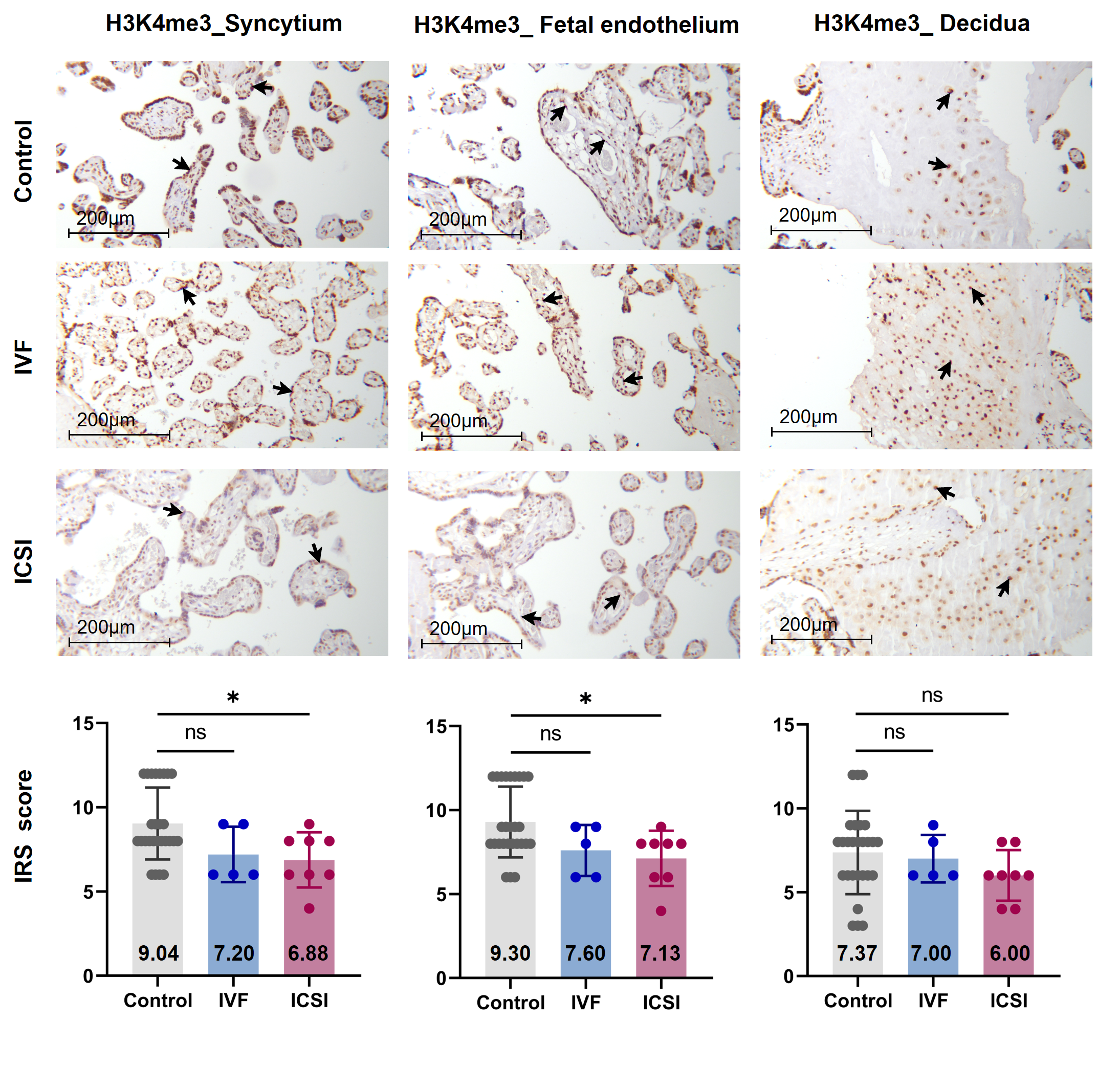
Figure 1. Global H3K4me3 was reduced in ICSI-derived placentas. Compared with natural conception placentas, ICSI placentas showed lower levels of H3K4me3 in syncytium and fetal endothelium, while IVF placentas showed no significant difference. Boxplots showing the IRS distribution among natural conception (n = 27), IVF (n = 5), ICSI (n = 8) groups along with significance levels indicated as P-value of the Mann-Whitney U-test. IRS = staining intensity × percentage of positive cells. Data were reported as mean ± SD. The arrows in the first, second, and third columns from left respectively pointed towards syncytial trophoblast cells, foetal endothelial cells and extravillous trophoblast cells. The numbers in bar charts represented the mean value of the corresponding IRS. H3K4me3, tri-methylated histone H3 lysine-4; IVF, in vitro fertilization; ICSI, intracytoplasmic sperm injection; IRS, immunoreactivity score; ns, non-significant. * P < 0.05.
3. The ICSI-Boys Present More Genes with Differentially Enriched H3K4me3 (deH3K4me3) Than In Vitro Fertilization (IVF)-Boys, ICSI-Girls, and IVF-Girls
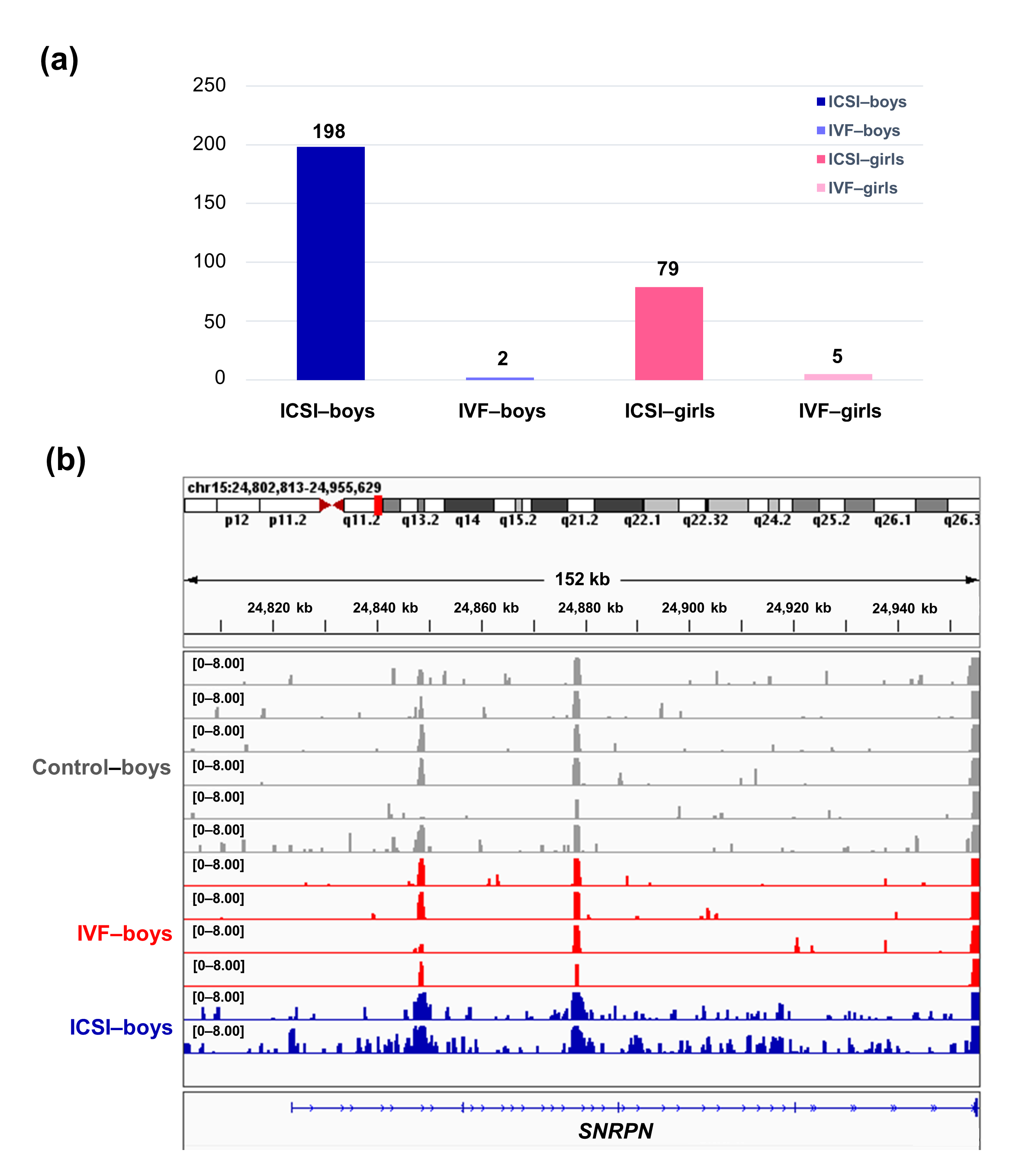
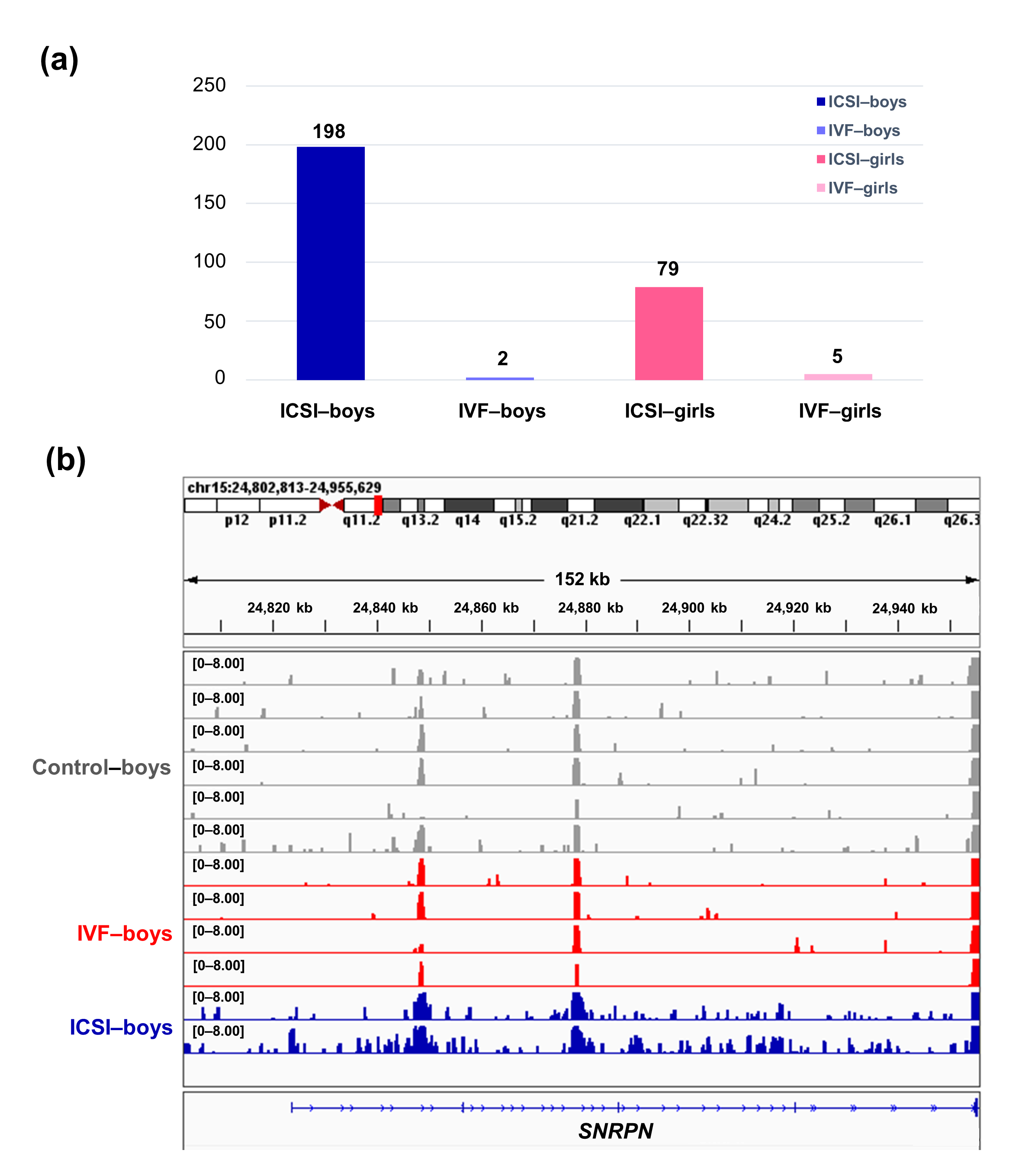
To further compare the sex-stratified H3K4me3 levels in newborn CBMC from natural conception and ARTs, H3K4me3 ChIP-seq data was taken from NCBI Gene Expression Omnibus (GEO, http://www.ncbi.nlm.nih.gov/geo/). When compared with the CBMC from same-gender naturally-conceived-children, the CBMC from ICSI-boys presented more genes with differentially enriched H3K4me3 (deH3K4me3) (n = 198, |log fold change (FC)| >1 and false discovery rate (FDR) < 0.05) than those from ICSI-girls (n = 79, |log FC| >1 and FDR < 0.05), IVF-girls (n = 5, |log FC| >1 and FDR < 0.05), and IVF-boys (n = 2, |log FC| >1 and FDR < 0.05) (Figure 2a). Likewise, Ghosh and colleagues [29] revealed that ART boys were more susceptible to ART-treatment-associated global DNA dys-methylation. Another study also revealed male offspring presented more robust responses and neurobehavior alterations to maternal inflammation [30]. One possible explanation is the sex-specific differences in the sensitivity to external exposure. But further studies are encouraged to make an intensive study and elucidate the underlying mechanisms.
We found ICSI-boys showed three imprinted genes with deH3K4me3 (i.e., Small Nuclear Ribonucleoprotein Polypeptide N [SNRPN], ZFP90 Zinc Finger Protein [ZFP90], and DiGeorge Syndrome Critical Region Gene 6 [DGCR6]) and ICSI-girls showed one imprinted gene with deH3K4me3 (i.e., HNF1 Homeobox A [HNF1A]) at promoter. The H3K4me3 enrichment of imprinted gene SNRPN from naturally-conceived-boys, IVF-boys, and ICSI-boys was shown in Figure 2b. Similarly, Choux and colleagues [31] checked five kinds of histone modifications in placentas from natural conception and IVF/ICSI, found the permissive marker H3K4me2 enrichment at the differentially methylated regions of H19/IGF2 and KCNQ1OT1 was significantly higher in IVF/ICSI group than those in natural conception group, and revealed that the epigenetic changes of imprinted genes at birth might be an important developmental event caused by ART manipulations. Rivera and colleagues [32] found that even the most basic manipulation (i.e., embryo transfer) could contribute to the mis-expression of imprinted genes during post-implantation development. While the in vitro culture followed by embryo transfer could make the situation worse, specifically by, increasing numbers of aberrant imprinted genes in mouse embryos, yolk sacs, and placentas, with even some fetuses exhibiting aberrant imprinted genes. The disruption of SNRPN has been revealed to play a role in AS and PWS, which are the major imprinting disorders in ART children [7][33].
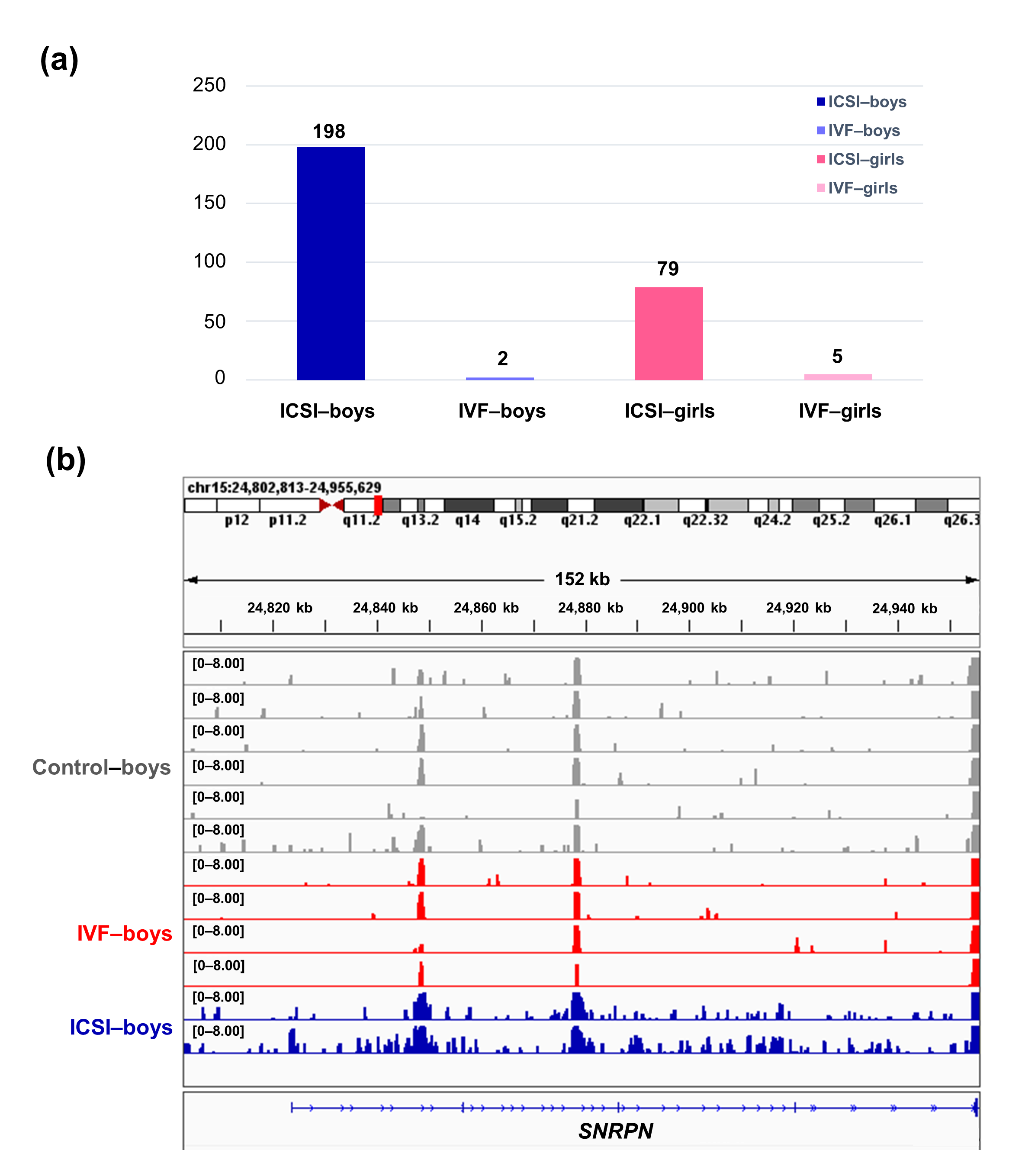 Figure 2. The ICSI-boys presented more genes with deH3K4me3 than IVF-boys, ICSI-girls, and IVF-girls. (a) The bar chart showed the comparison results of each gene promoter H3K4me3 read counts between natural conception-boys (n = 6) vs ICSI-boys (n = 2), natural conception-boys (n = 6) vs IVF-boys (n = 4), natural conception-girls (n = 6) vs ICSI-girls (n = 4), and natural conception-girls (n = 6) vs IVF-girls (n = 4). The comparison was performed via R package 'edgeR'. The numbers above the columns were the numbers of genes with significant H3K4me3 alteration in promoter region (|log FC| >1 and FDR < 0.05). (b) Genome browser snapshots of H3K4me3 ChIP-seq data at SNRPN loci in naturally-conceived-boys, IVF-boys, and ICSI-boys. H3K4me3 data showed the log2 enrichment ratio between H3K4me3 ChIP and the input. The data range was set at 0—8.00. deH3K4me3, differentially enriched H3K4me3; FC, fold change; FDR, false discovery rate; SNRPN, small nuclear ribonucleoprotein polypeptide N.
Figure 2. The ICSI-boys presented more genes with deH3K4me3 than IVF-boys, ICSI-girls, and IVF-girls. (a) The bar chart showed the comparison results of each gene promoter H3K4me3 read counts between natural conception-boys (n = 6) vs ICSI-boys (n = 2), natural conception-boys (n = 6) vs IVF-boys (n = 4), natural conception-girls (n = 6) vs ICSI-girls (n = 4), and natural conception-girls (n = 6) vs IVF-girls (n = 4). The comparison was performed via R package 'edgeR'. The numbers above the columns were the numbers of genes with significant H3K4me3 alteration in promoter region (|log FC| >1 and FDR < 0.05). (b) Genome browser snapshots of H3K4me3 ChIP-seq data at SNRPN loci in naturally-conceived-boys, IVF-boys, and ICSI-boys. H3K4me3 data showed the log2 enrichment ratio between H3K4me3 ChIP and the input. The data range was set at 0—8.00. deH3K4me3, differentially enriched H3K4me3; FC, fold change; FDR, false discovery rate; SNRPN, small nuclear ribonucleoprotein polypeptide N.
4. RNA Polymerase II Subunit A (Polr2A) and Lysine Demethylase 5A (KDM5A) Are the Regulators of H3K4me3
Typically, enzymes that catalyze H3K4me3 include both histone lysine methyltransferases (known as 'writers') and demethylases (known as 'erasers'). These enzymes mediate the dynamic regulation of H3K4me3 [34]. Other proteins/molecules, such as Pygopus 2 (Pygo2)[35], Polycomb group ring finger 6 (PCGF6) [44] and Interleukin-13 (IL-13) [36], have also been found to be involved in H3K4me3 regulation. In the current study, through transcription factor (TF) analysis by 'RcisTarget' package and literature retrieval, we found RNA polymerase II subunit A (Polr2A) and lysine demethylase 5A (KDM5A) were potentially involved in regulating H3K4me3 modification. We conducted IHC staining and IRS evaluation for Polr2A and KDM5A in placental tissues from natural conception, IVF, and ICSI. Polr2A expression showed no significant difference between groups (P > 0.05). KDM5A expression (Figure 3) was increased in the syncytium (P < 0.001), fetal endothelium (P = 0.001), and decidua (P = 0.048) of ICSI placentas than those in naturally-conceived-placentas. KDM5A expression showed no significant difference between natural conception and IVF groups (P > 0.05). We transfected HTR-8/SVneo cells with siRNA targeting Polr2A (si-Polr2A), KDM5A (si-KDM5A), and non-targeting control siRNA (si-NT). Western blot analysis (Figure 4a—c) and Immunocytochemistry (ICC) staining (Figure 4d) revealed that knockdown of Polr2A and KDM5A respectively resulted in down- and up-regulation of H3K4me3.
Polr2A is the largest subunit of RNA polymerase II (Pol II), and it plays a fundamental part in the enzymatic activity of Pol II [37]. A previous study revealed that in oxidative stress conditions, Polr2A could be regulated by von Hippel–Lindau protein (pVHL) and proline hydroxylases (PHDs) [46] (known as 'oxygen sensors' [38]. Treatment of 786-O cells with hydrogen peroxide (H2O2) resulted in significant induction of phosphorylated Polr2A [37]. In addition, intermittent hypoxia and UV irradiation have also been shown to be able to induce Polr2A ubiquitylation and degradation [39][40]. As mentioned above, Polr2A is a fundamental part of Pol II. And a previous study revealed that H3K4me3-associated active interacting domains were mostly embedded in Pol II-associated transcriptional interacting domains [41]. Herein, our study provided the first evidence that Polr2A positively regulated H3K4me3 enrichment, revealing Polr2A might play a role in promoting a more permissive chromatin conformation [42].
KDM5A is a histone lysine demethylase (KDM) and is described as an 'oxygen sensor', because the activity of KDM5A is oxygen-sensitive in various cellular systems [43]. In depleted oxygen conditions, KDMs' activity is inhibited, and as compensation, the expressions of KDMs are commensurately increased[44] . A study of HepG2 hepatoma cells reported that mRNA and protein levels of KDM5B were higher following exposure to low and severely low oxygen tensions [45]. However, in another study (on Beas-2B cells), low-oxygen treatment was not found to cause any significant change of KDM5A in both mRNA and protein levels . The variability of the findings in these studies may, however, reflect different experimental conditions and cell type specificities. Notably, the increased histone trimethylation under low oxygen tensions mainly resulted from attenuated catalytic activities of histone demethylases and not from altered abundances of histone demethylases[44]. KDM5A is able to catalyze the removal of all three methyl groups from H3K4 lysine residue and is essential for early embryo development . Knockdown of KDM5A in HeLa cells is stated to contribute to the global increase of H3K4me3 [46]. And our knockdown experiment in HTR-8/SVneo cells further verified that KDM5A is a negative regulator of H3K4me3. In the IHC staining, KDM5A also showed an opposite expression trend compared to H3K4me3.
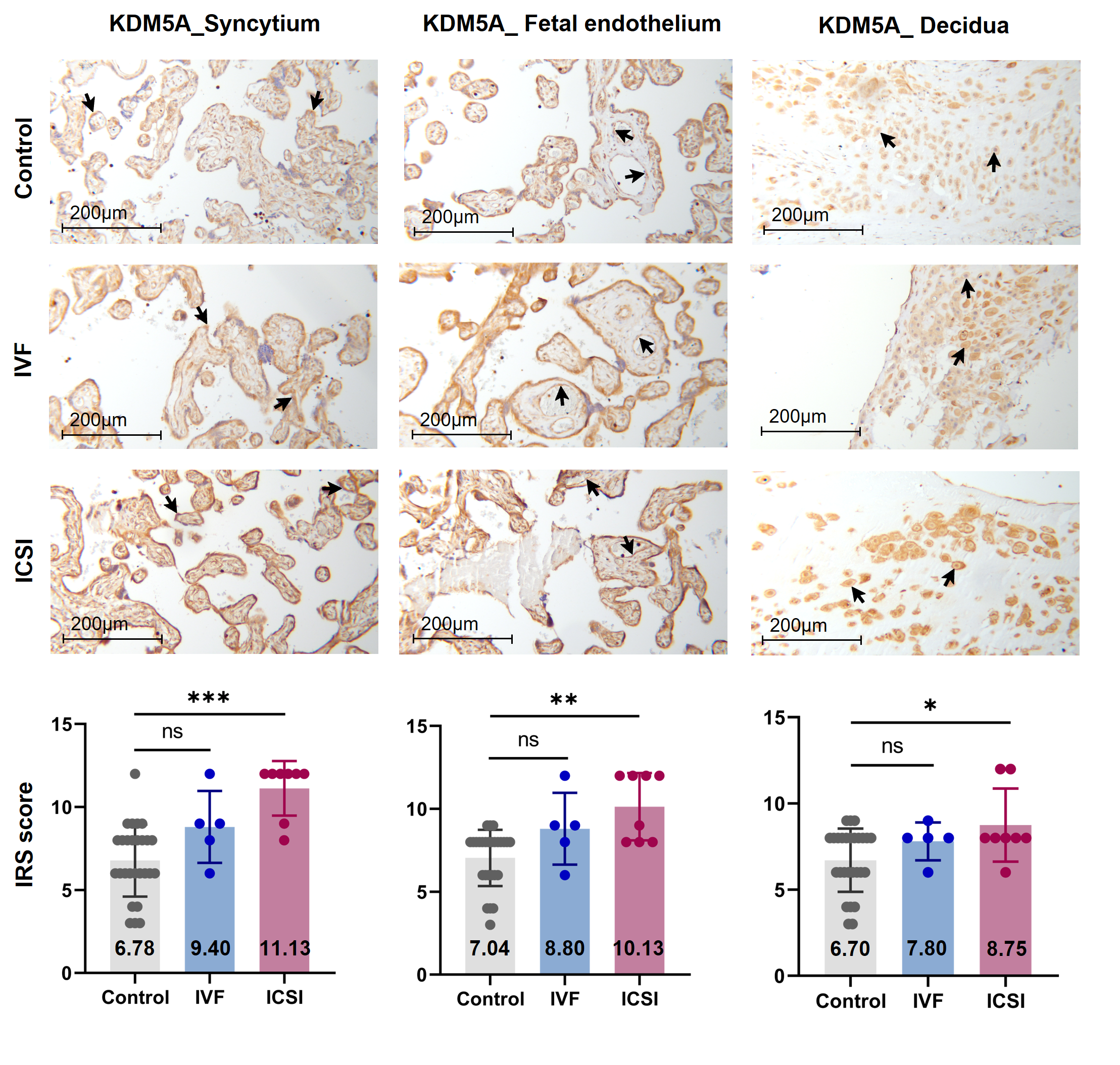 Figure 3. KDM5A expression was increased in ICSI-derived placentas. Compared with natural conception placentas, ICSI placentas showed higher levels of KDM5A in syncytium, fetal endothelium, and decidua, while IVF placentas showed no significant difference. Boxplots showing the IRS distribution among natural conception (n = 27), IVF (n = 5), ICSI (n = 8) groups along with significance levels indicated as P-value of the Mann-Whitney U-test. IRS = staining intensity × percentage of positive cells. Data were reported as mean ± SD. The arrows in the first, second, and third columns from left respectively pointed towards syncytial trophoblast cells, foetal endothelial cells and extravillous trophoblast cells. The numbers in bar charts represented the mean value of the corresponding IRS. KDM5A, lysine demethylase 5A. * P < 0.05; ** P < 0.01; *** P < 0.001.
Figure 3. KDM5A expression was increased in ICSI-derived placentas. Compared with natural conception placentas, ICSI placentas showed higher levels of KDM5A in syncytium, fetal endothelium, and decidua, while IVF placentas showed no significant difference. Boxplots showing the IRS distribution among natural conception (n = 27), IVF (n = 5), ICSI (n = 8) groups along with significance levels indicated as P-value of the Mann-Whitney U-test. IRS = staining intensity × percentage of positive cells. Data were reported as mean ± SD. The arrows in the first, second, and third columns from left respectively pointed towards syncytial trophoblast cells, foetal endothelial cells and extravillous trophoblast cells. The numbers in bar charts represented the mean value of the corresponding IRS. KDM5A, lysine demethylase 5A. * P < 0.05; ** P < 0.01; *** P < 0.001.
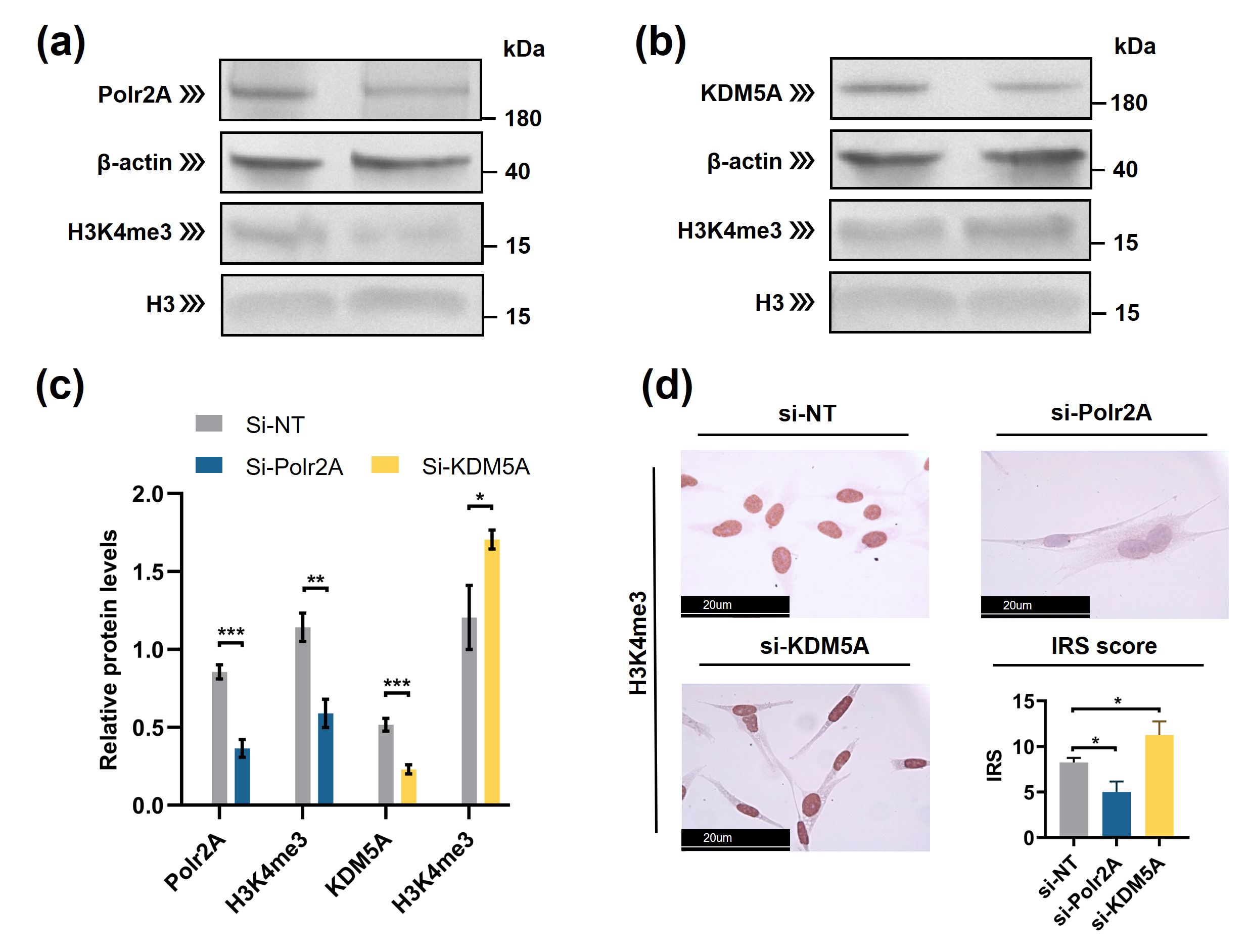 Figure 4. Polr2A and KDM5A regulated global levels of H3K4me3 in HTR-8/SVneo cells. (a—c) Western blot analysis and (d) immunocytochemistry showed that, compared with si-NT-cells, si-Polr2A-cells showed lower levels of H3K4me3 and si-KDM5A-cells showed higher levels of H3K4me3. In the boxplots, protein levels of Polr2A and KDM5A expressed relative to β-actin levels, protein levels of H3K4me3 expressed relative to histone H3 levels, IRS = staining intensity × percentage of positive cells. Three independent experiments were performed. Data were reported as mean ± SD. Independent samples t-test was used for statistical analysis. H3, Histone H3; si-NT, non-targeting control small interfering RNA; si-Polr2A, siRNA targeting Polr2A; si-KDM5A, siRNA targeting KDM5A. * P < 0.05; ** P < 0.01; *** P < 0.001.
Figure 4. Polr2A and KDM5A regulated global levels of H3K4me3 in HTR-8/SVneo cells. (a—c) Western blot analysis and (d) immunocytochemistry showed that, compared with si-NT-cells, si-Polr2A-cells showed lower levels of H3K4me3 and si-KDM5A-cells showed higher levels of H3K4me3. In the boxplots, protein levels of Polr2A and KDM5A expressed relative to β-actin levels, protein levels of H3K4me3 expressed relative to histone H3 levels, IRS = staining intensity × percentage of positive cells. Three independent experiments were performed. Data were reported as mean ± SD. Independent samples t-test was used for statistical analysis. H3, Histone H3; si-NT, non-targeting control small interfering RNA; si-Polr2A, siRNA targeting Polr2A; si-KDM5A, siRNA targeting KDM5A. * P < 0.05; ** P < 0.01; *** P < 0.001.
5. Varying Oxygen Conditions Regulate Protein Levels of H3K4me3
Next, to investigate the influence of oxygen tensions on global levels of H3K4me3 and its regulators, we cultured HTR-8/SVneo cells in different oxygen conditions. Western blot analysis (Figure 5) revealed after low-oxygen culture (1% and 5% O2), global levels of H3K4me3 were significantly higher than those after atmospheric oxygen culture. Besides, after intermittent hyperoxia exposure, global H3K4me3 levels were significantly different from those after persistent atmospheric culture and low-oxygen culture (5% O2). Our findings agree with previous studies [47][48][49], which have reported that in low oxygen tensions (1% O2 and less than 0.5% O2), global levels of H3K4me3 increased in multiple human cell lines (e.g., HeLa cells, HFF cells, A549 cells, Beas-2B cells, and hADSC Cells). Intermittent hyperoxia exposure may also contribute to H3K4me3 alteration[50]. Further, we hypothesize that the oxygen tensions and intermittent hyperoxia exposure-associated H3K4me3 alteration in our cell culture system, may have been due to increased production of reactive oxygen species (ROS); it is known that, higher O2 tension and re-oxygenation are both accompanied by increased generation of ROS[51][52]. Unlike the in vivo system, in vitro culturing conditions lack naturally effective antioxidant systems[53]. Excessive ROS produced in the impaired cellular antioxidant systems may further contribute to redox imbalance [54], influencing redox-sensitive TFs and critical enzymes associated with histone modifications[53]. One previous study has also proposed that H3K4me3 is redox-regulated, and H2O2 (a ROS generator) treatment in HeLa cells results in decreased levels of H3K4me3[55] Currently, in human embryo culturing systems, it is common to supplement antioxidants, yet how to maintain a prooxidant-antioxidant equilibrium still deserves further investigation.
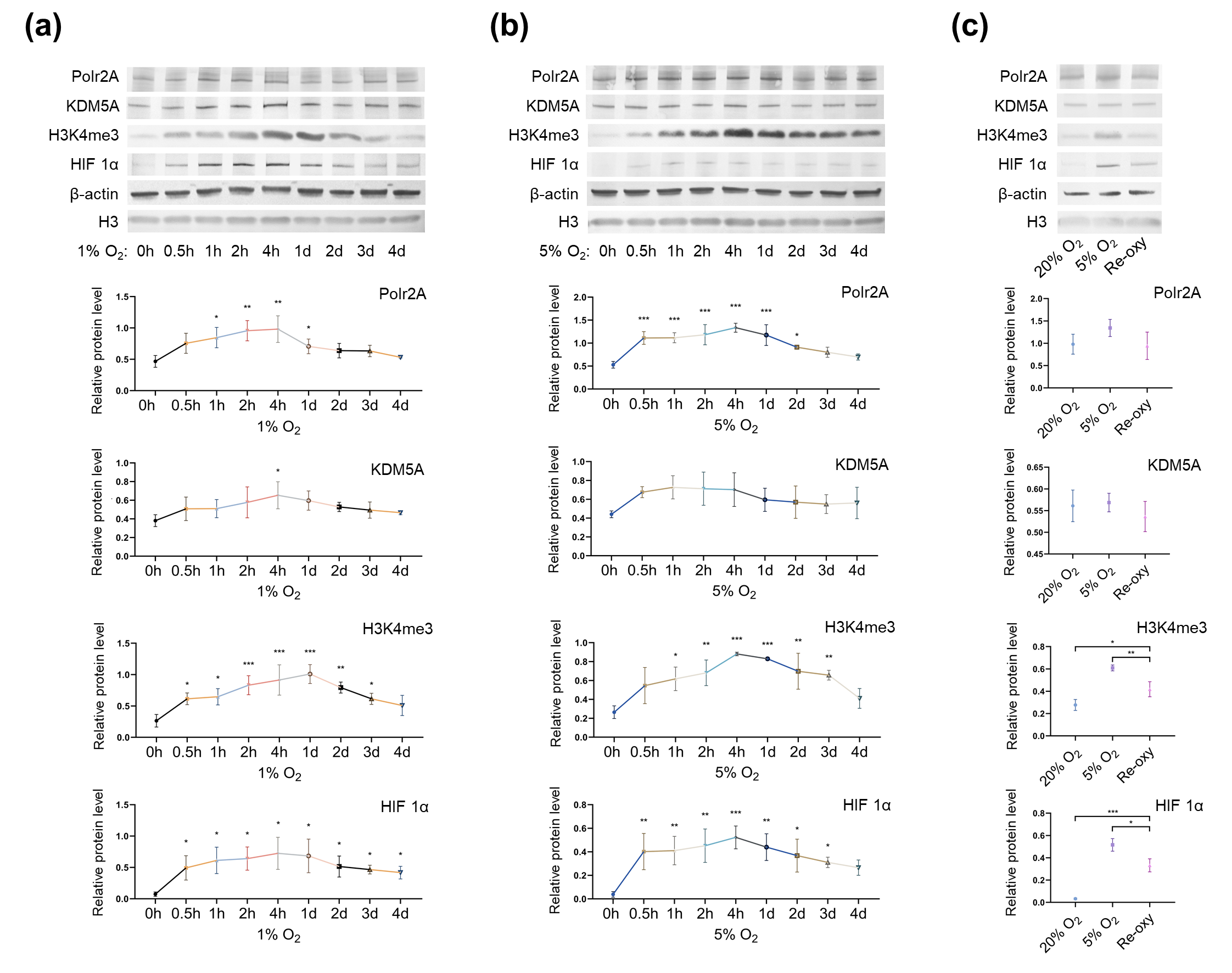 Figure 5. Protein levels of Polr2A, KDM5A, and H3K4me3 in varying oxygen conditions. After low-oxygen culture at 1% (a) and 5% O2 (b), protein levels of Polr2A, H3K4me3, and HIF 1α were higher than atmospheric oxygen culture. After intermittent hyperoxia exposure (c), protein levels of H3K4me3 and HIF 1α were different from those after persistent atmospheric oxygen culture or persistent low-oxygen culture (5% O2). In the boxplots, protein levels of Polr2A, KDM5A, and HIF 1α expressed relative to β-actin levels, protein levels of H3K4me3 expressed relative to histone H3 levels. Three independent experiments were performed. Data were reported as mean ± SD. One-way analysis of variance with Dunnet's posthoc test and independent samples t-test were used for statistical analysis. HIF 1α was used as the indicator for hypoxia. HIF 1α, hypoxia-inducible factor 1α. 'Re-oxy' referred to intermittent hyperoxia exposure (5% O2 8 hours + 20% O2 16 hours + 5% O2 8 hours). * P < 0.05; ** P < 0.01; *** P < 0.001.
Figure 5. Protein levels of Polr2A, KDM5A, and H3K4me3 in varying oxygen conditions. After low-oxygen culture at 1% (a) and 5% O2 (b), protein levels of Polr2A, H3K4me3, and HIF 1α were higher than atmospheric oxygen culture. After intermittent hyperoxia exposure (c), protein levels of H3K4me3 and HIF 1α were different from those after persistent atmospheric oxygen culture or persistent low-oxygen culture (5% O2). In the boxplots, protein levels of Polr2A, KDM5A, and HIF 1α expressed relative to β-actin levels, protein levels of H3K4me3 expressed relative to histone H3 levels. Three independent experiments were performed. Data were reported as mean ± SD. One-way analysis of variance with Dunnet's posthoc test and independent samples t-test were used for statistical analysis. HIF 1α was used as the indicator for hypoxia. HIF 1α, hypoxia-inducible factor 1α. 'Re-oxy' referred to intermittent hyperoxia exposure (5% O2 8 hours + 20% O2 16 hours + 5% O2 8 hours). * P < 0.05; ** P < 0.01; *** P < 0.001.
6. Conclusions
Given the small sample size, our power to make a definitive conclusion is limited. Nevertheless, some interesting observations still merit attention: when compared with the naturally-conceived-group, placenta and newborn CBMC from ICSI, but not IVF, group showed H3K4me3 alteration. Besides, ICSI-boys presented more genes with deH3K4me3 than ICSI-girls. Varying oxygen conditions, Polr2A and KDM5A impacted H3K4me3 levels. Considering the ICSI is usually prepared for male oligospermia or asthenospermia or abnormal fertilization in previous conventional IVF, the alteration of H3K4me3 in ICSI group may not only derive from ICSI technology per ser but also from the couple's infertility background.
References
- Jagannathan, L.; Cuddapah, S.; Costa, M. Oxidative Stress under Ambient and Physiological Oxygen Tension in Tissue Culture. Curr. Pharmacol. Rep. 2016, 2, 64–72. [Google Scholar] [CrossRef] [PubMed]
- Gosden, R.; Trasler, J.; Lucifero, D.; Faddy, M. Rare congenital disorders, imprinted genes, and assisted reproductive technology. Lancet 2003, 361, 1975–1977. [Google Scholar] [CrossRef]
- Liu, Y.; Gu, Y.; Su, M.; Liu, H.; Zhang, S.; Zhang, Y. An analysis about heterogeneity among cancers based on the DNA methylation patterns. BMC Cancer 2019, 19, 1–15. [Google Scholar] [CrossRef]
- Rexhaj, E.; Paoloni-Giacobino, A.; Rimoldi, S.F.; Fuster, D.G.; Anderegg, M.; Somm, E.; Bouillet, E.; Allemann, Y.; Sartori, C.; Scherrer, U. Mice generated by in vitro fertilization exhibit vascular dysfunction and shortened life span. J. Clin. Investig. 2013, 123, 5052–5060. [Google Scholar] [CrossRef] [PubMed]
- Rexhaj, E.; Bloch, J.; Jayet, P.Y.; Rimoldi, S.F.; Dessen, P.; Mathieu, C.; Tolsa, J.F.; Nicod, P.; Scherrer, U.; Sartori, C. Fetal programming of pulmonary vascular dysfunction in mice: Role of epigenetic mechanisms. Am. J. Physiol. Heart Circ. Physiol. 2011, 301, H247–H252. [Google Scholar] [CrossRef] [PubMed]
- Market-Velker, B.A.; Fernandes, A.D.; Mann, M.R.W. Side-by-side comparison of five commercial media systems in a mouse model: Suboptimal in vitro culture interferes with imprint maintenance. Biol. Reprod. 2010, 83, 938–950. [Google Scholar] [CrossRef] [PubMed]
- Uk, A.; Collardeau-Frachon, S.; Scanvion, Q.; Michon, L.; Amar, E. Assisted Reproductive Technologies and imprinting disorders: Results of a study from a French congenital malformations registry. Eur. J. Med. Genet. 2018, 61, 518–523. [Google Scholar] [CrossRef] [PubMed]
- Lazaraviciute, G.; Kauser, M.; Bhattacharya, S.; Haggarty, P.; Bhattacharya, S. A systematic review and meta-analysis of DNA methylation levels and imprinting disorders in children conceived by IVF/ICSI compared with children conceived spontaneously. Hum. Reprod. Update 2014, 20, 840–852. [Google Scholar] [CrossRef]
- Ghosh, J.; Coutifaris, C.; Sapienza, C.; Mainigi, M. Global DNA methylation levels are altered by modifiable clinical manipulations in assisted reproductive technologies. Clin. Epigenetics 2017, 9, 1–10. [Google Scholar] [CrossRef]
- Kobayashi, H.; Hiura, H.; John, R.M.; Sato, A.; Otsu, E.; Kobayashi, N.; Suzuki, R.; Suzuki, F.; Hayashi, C.; Utsunomiya, T.; et al. DNA methylation errors at imprinted loci after assisted conception originate in the parental sperm. Eur. J. Hum. Genet. 2009, 17, 1582–1591. [Google Scholar] [CrossRef]
- Fernández-Gonzalez, R.; Ramirez, M.A.; Bilbao, A.; De Fonseca, F.R.; Gutiérrez-Adán, A. Suboptimal in vitro culture conditions: An epigenetic origin of long-term health effects. Mol. Reprod. Dev. 2007, 74, 1149–1156. [Google Scholar] [CrossRef]
- De waal, E.; Yamazaki, Y.; Ingale, P.; Bartolomei, M.S.; Yanagimachi, R.; McCarrey, J.R. Gonadotropin stimulation contributes to an increased incidence of epimutations in ICSI-derived mice. Hum. Mol. Genet. 2012, 21, 4460–4472. [Google Scholar] [CrossRef] [PubMed]
- Turner, B.M. Epigenetic responses to environmental change and their evolutionary implications. Philos. Trans. R. Soc. B Biol. Sci. 2009, 364, 3403–3418. [Google Scholar] [CrossRef] [PubMed]
- Eastman, A.J.; Xu, J.; Bermik, J.; Potchen, N.; den Dekker, A.; Neal, L.M.; Zhao, G.; Malachowski, A.; Schaller, M.; Kunkel, S.; et al. Epigenetic stabilization of DC and DC precursor classical activation by TNFα contributes to protective T cell polarization. Sci. Adv. 2019, 5, eaaw9051. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Q.; Bai, B.; Mei, X.; Wan, C.; Cao, H.; Li, D.; Wang, S.; Zhang, M.; Wang, Z.; Wu, J.; et al. Elevated H3K79 homocysteinylation causes abnormal gene expression during neural development and subsequent neural tube defects. Nat. Commun. 2018, 9, 1–16. [Google Scholar] [CrossRef] [PubMed]
- Huang, X.; Gao, X.; Li, W.; Jiang, S.; Li, R.; Hong, H.; Zhao, C.; Zhou, P.; Chen, H.; Bo, X.; et al. Stable H3K4me3 is associated with transcription initiation during early embryo development. Bioinformatics 2019, 35, 3931–3936. [Google Scholar] [CrossRef]
- Liu, M.; Chen, B.; Pei, L.; Zhang, Q.; Zou, Y.; Xiao, H.; Zhou, J.; Chen, L.; Wang, H. Decreased H3K9ac level of StAR mediated testicular dysplasia induced by prenatal dexamethasone exposure in male offspring rats. Toxicology 2018, 408, 1–10. [Google Scholar] [CrossRef]
- Xiao, H.; Wen, Y.; Pan, Z.; Shangguan, Y.; Qin, J.; Tan, Y.; Jiang, H.; Li, B.; Zhang, Q.; Chen, L.; et al. Increased H3K27ac level of ACE mediates the intergenerational effect of low peak bone mass induced by prenatal dexamethasone exposure in male offspring rats. Cell Death Dis. 2018, 9, 1–14. [Google Scholar] [CrossRef]
- Creyghton, M.P.; Cheng, A.W.; Welstead, G.G.; Kooistra, T.; Carey, B.W.; Steine, E.J.; Hanna, J.; Lodato, M.A.; Frampton, G.M.; Sharp, P.A.; et al. Histone H3K27ac separates active from poised enhancers and predicts developmental state. Proc. Natl. Acad. Sci. USA 2010, 107, 21931–21936. [Google Scholar] [CrossRef]
- Okae, H.; Toh, H.; Sato, T.; Hiura, H.; Takahashi, S.; Shirane, K.; Kabayama, Y.; Suyama, M.; Sasaki, H.; Arima, T. Derivation of Human Trophoblast Stem Cells. Cell Stem Cell 2018, 22, 50–63.e6. [Google Scholar] [CrossRef]
- Gaspar, R.C.; Arnold, D.R.; Corrêa, C.A.P.; da Rocha, C.V.; Penteado, J.C.T.; del Collado, M.; Vantini, R.; Garcia, J.M.; Lopes, F.L. Oxygen tension affects histone remodeling of invitro-produced embryos in a bovine model. Theriogenology 2015, 83, 1408–1415. [Google Scholar] [CrossRef]
- Belli, M.; Antonouli, S.; Palmerini, M.G.; Bianchi, S.; Bernardi, S.; Khalili, M.A.; Donfrancesco, O.; Nottola, S.A.; Macchiarelli, G. The effect of low and ultra-low oxygen tensions on mammalian embryo culture and development in experimental and clinical IVF. Syst. Biol. Reprod. Med. 2020, 66, 229–235. [Google Scholar] [CrossRef]
- Bontekoe, S.; Mantikou, E.; van Wely, M.; Seshadri, S.; Repping, S.; Mastenbroek, S. Low oxygen concentrations for embryo culture in assisted reproductive technologies. Cochrane Database Syst. Rev. 2012, 7, CD008950. [Google Scholar] [CrossRef] [PubMed]
- Guarneri, C.; Restelli, L.; Mangiarini, A.; Ferrari, S.; Somigliana, E.; Paffoni, A. Can we use incubators with atmospheric oxygen tension in the first phase of in vitro fertilization? A retrospective analysis. J. Assist. Reprod. Genet. 2015, 32, 77–82. [Google Scholar] [CrossRef] [PubMed]
- Chen, W.; Peng, Y.; Ma, X.; Kong, S.; Tan, S.; Wei, Y.; Zhao, Y.; Zhang, W.; Wang, Y.; Yan, L.; et al. Integrated multi-omics reveal epigenomic disturbance of assisted reproductive technologies in human offspring. EBioMedicine 2020, 61, 103076. [Google Scholar] [CrossRef] [PubMed]
- Elliott, G.; Hong, C.; Xing, X.; Zhou, X.; Li, D.; Coarfa, C.; Bell, R.J.A.; Maire, C.L.; Ligon, K.L.; Sigaroudinia, M.; et al. Intermediate DNA methylation is a conserved signature of genome regulation. Nat. Commun. 2015, 6, 7363. [Google Scholar] [CrossRef]
- Kim, T.H.; Barrera, L.O.; Zheng, M.; Qu, C.; Singer, M.A.; Richmond, T.A.; Wu, Y.; Green, R.D.; Ren, B. A high-resolution map of active promoters in the human genome. Nature 2005, 436, 876–880. [Google Scholar] [CrossRef] [PubMed]
- Niu, Y.; Desmarais, T.L.; Tong, Z.; Yao, Y.; Costa, M. Oxidative stress alters global histone modification and DNA methylation. Free Radic. Biol. Med. 2015, 82, 22–28. [Google Scholar] [CrossRef]
- Guo, X.Y.; Liu, X.M.; Jin, L.; Wang, T.T.; Ullah, K.; Sheng, J.Z.; Huang, H.F. Cardiovascular and metabolic profiles of offspring conceived by assisted reproductive technologies: A systematic review and meta-analysis. Fertil. Steril. 2017, 107, 622–631.e5. [Google Scholar] [CrossRef]
- Graf, A.E.; Lallier, S.W.; Waidyaratne, G.; Thompson, M.D.; Tipple, T.E.; Hester, M.E.; Trask, A.J.; Rogers, L.K. Maternal high fat diet exposure is associated with increased hepcidin levels, decreased myelination, and neurobehavioral changes in male offspring. Brain. Behav. Immun. 2016, 58, 369–378. [Google Scholar] [CrossRef]
- Choux, C.; Petazzi, P.; Sanchez-Delgado, M.; Hernandez Mora, J.R.; Monteagudo, A.; Sagot, P.; Monk, D.; Fauque, P. The hypomethylation of imprinted genes in IVF/ICSI placenta samples is associated with concomitant changes in histone modifications. Epigenetics 2020, 15, 1386–1395. [Google Scholar] [CrossRef]
- Rivera, R.M.; Stein, P.; Weaver, J.R.; Mager, J.; Schultz, R.M.; Bartolomei, M.S. Manipulations of mouse embryos prior to implantation result in aberrant expression of imprinted genes on day 9.5 of development. Hum. Mol. Genet. 2008, 17, 1–14. [Google Scholar] [CrossRef]
- White, H.E.; Hall, V.J.; Cross, N.C.P. Methylation-sensitive high-resolution melting-curve analysis of the SNRPN gene as a diagnostic screen for Prader-Willi and Angelman syndromes. Clin. Chem. 2007, 53, 1960–1962. [Google Scholar] [CrossRef] [PubMed]
- Araki, Y.; Aizaki, Y.; Sato, K.; Oda, H.; Kurokawa, R.; Mimura, T. Altered gene expression profiles of histone lysine methyltransferases and demethylases in rheumatoid arthritis synovial fibroblasts. Clin. Exp. Rheumatol. 2018, 36, 314–316. [Google Scholar]
- Zhou, C.; Zhang, Y.; Dai, J.; Zhou, M.; Liu, M.; Wang, Y.; Chen, X.Z.; Tang, J. Pygo2 functions as a prognostic factor for glioma due to its up-regulation of H3K4me3 and promotion of MLL1/MLL2 complex recruitment. Sci. Rep. 2016, 6, 22066. [Google Scholar] [CrossRef] [PubMed]
- Yu, L.; Li, N.; Zhang, J.; Jiang, Y. IL-13 regulates human nasal epithelial cell differentiation via H3K4me3 modification. J. Inflamm. Res. 2017, 10, 181–188. [Google Scholar] [CrossRef] [PubMed]
- Mikhaylova, O.; Ignacak, M.L.; Barankiewicz, T.J.; Harbaugh, S.V.; Yi, Y.; Maxwell, P.H.; Schneider, M.; Van Geyte, K.; Carmeliet, P.; Revelo, M.P.; et al. The von Hippel-Lindau Tumor Suppressor Protein and Egl-9-Type Proline Hydroxylases Regulate the Large Subunit of RNA Polymerase II in Response to Oxidative Stress. Mol. Cell. Biol. 2008, 28, 2701–2717. [Google Scholar] [CrossRef]
- Li, X.; Wang, T.; Yin, S.; Zhang, G.; Cao, Q.; Wen, X.; Zhang, H.; Wang, D.; Zhu, W. The improved energy metabolism and blood oxygen-carrying capacity for pufferfish, Takifugu fasciatus, against acute hypoxia under the regulation of oxygen sensors. Fish Physiol. Biochem. 2019, 45, 323–340. [Google Scholar] [CrossRef]
- Ignacak, M.L.; Harbaugh, S.V.; Dayyat, E.; Row, B.W.; Gozal, D.; Czyzyk-Krzeska, M.F. Intermittent hypoxia regulates RNA polymerase II in hippocampus and prefrontal cortex. Neuroscience 2009, 158, 1436–1445. [Google Scholar] [CrossRef] [PubMed]
- Tufegdžić Vidaković, A.; Mitter, R.; Kelly, G.P.; Neumann, M.; Harreman, M.; Rodríguez-Martínez, M.; Herlihy, A.; Weems, J.C.; Boeing, S.; Encheva, V.; et al. Regulation of the RNAPII Pool Is Integral to the DNA Damage Response. Cell 2020, 180, 1245–1261.e21. [Google Scholar] [CrossRef]
- Zhao, L.; Wang, S.; Cao, Z.; Ouyang, W.; Zhang, Q.; Xie, L.; Zheng, R.; Guo, M.; Ma, M.; Hu, Z.; et al. Chromatin loops associated with active genes and heterochromatin shape rice genome architecture for transcriptional regulation. Nat. Commun. 2019, 10, 3640. [Google Scholar] [CrossRef]
- Rea, S.; Eisenhaber, F.; O’Carroll, D.; Strahl, B.D.; Sun, Z.W.; Schmid, M.; Opravil, S.; Mechtier, K.; Ponting, C.P.; Allis, C.D.; et al. Regulation of chromatin structure by site-specific histone H3 methyltransferases. Nature 2000, 406, 593–599. [Google Scholar] [CrossRef]
- Gallipoli, P.; Huntly, B.J.P. Histone modifiers are oxygen sensors. Science 2019, 363, 1148–1149. [Google Scholar] [CrossRef] [PubMed]
- Lee, S.; Lee, J.; Chae, S.; Moon, Y.; Lee, H.Y.; Park, B.; Yang, E.G.; Hwang, D.; Park, H.; Multi-dimensional histone methylations for coordinated regulation of gene expression under hypoxia.. Lee, S.; Lee, J.; Chae, S.; Moon, Y.; Lee, H.Y.; Park, B.; Yang, E.G.; Hwang, D.; Park, H. 2017, 45, 11643–11657.
- Xia, O.; Lemieux, M.E.; Li, W.; Carroll, J.S.; Brown, M.; Shirley Liu, X.; Kung, A.L. Integrative analysis of HIF binding and transactivation reveals its role in maintaining histone methylation homeostasis. Proc. Natl. Acad. Sci. USA 2009, 106, 4260–4265. [Google Scholar] [CrossRef]
- Dahl, J.A.; Jung, I.; Aanes, H.; Greggains, G.D.; Manaf, A.; Lerdrup, M.; Li, G.; Kuan, S.; Li, B.; Lee, A.Y.; et al. Broad histone H3K4me3 domains in mouse oocytes modulate maternal-to-zygotic transition. Nature 2016, 537, 548–552. [Google Scholar] [CrossRef]
- Batie, M.; Frost, J.; Frost, M.; Wilson, J.W.; Schofield, P.; Rocha, S. Hypoxia induces rapid changes to histone methylation and reprograms chromatin. Science 2019, 363, 1222–1226. [Google Scholar] [CrossRef]
- Lee, S.; Lee, J.; Chae, S.; Moon, Y.; Lee, H.Y.; Park, B.; Yang, E.G.; Hwang, D.; Park, H. Multi-dimensional histone methylations for coordinated regulation of gene expression under hypoxia. Nucleic Acids Res. 2017, 45, 11643–11657. [Google Scholar] [CrossRef]
- Zhou, X.; Sun, H.; Chen, H.; Zavadil, J.; Kluz, T.; Arita, A.; Costa, M. Hypoxia induces trimethylated H3 lysine 4 by inhibition of JARID1A demethylase. Cancer Res. 2010, 70, 4214–4221. [Google Scholar] [CrossRef]
- Yang, H.; Kuhn, C.; Kolben, T.; Ma, Z.; Lin, P.; Mahner, S.; Jeschke, U.; von Schönfeldt, V. Early life oxidative stress and long-lasting cardiovascular effects on offspring conceived by assisted reproductive technologies: A review. Int. J. Mol. Sci. 2020, 21, 5175. [Google Scholar] [CrossRef] [PubMed]
- Agarwal, A.; Said, T.M.; Bedaiwy, M.A.; Banerjee, J.; Alvarez, J.G. Oxidative stress in an assisted reproductive techniques setting. Fertil. Steril. 2006, 86, 503–512. [Google Scholar] [CrossRef]
- Sazontova, T.G.; Bolotova, A.V.; Bedareva, I.V.; Kostina, N.V.; Arkhipenko, Y.V. Adaptation to intermittent hypoxia/hyperoxia enhances efficiency of exercise training. In Intermittent Hypoxia and Human Diseases; Springer: London, UK, 2014; pp. 191–205. ISBN 9781447129066. [Google Scholar]
- Jagannathan, L.; Cuddapah, S.; Costa, M.; Oxidative Stress under Ambient and Physiological Oxygen Tension in Tissue Culture. Curr. Pharmacol. Rep. 2016, 2, 64–72.
- Parker, L.; Shaw, C.S.; Banting, L.; Levinger, I.; Hill, K.M.; McAinch, A.J.; Stepto, N.K.; cute low-volume high-intensity interval exercise and continuous moderate-intensity exercise elicit a similar improvement in 24-h glycemic control in overweight and obese adults.. Front. Physiol. 2017, 7, 661.
- Daphne Bazopoulou; Daniela Knoefler; Yongxin Zheng; Kathrin Ulrich; Bryndon J. Oleson; Lihan Xie; Minwook Kim; Anke Kaufmann; Young-Tae Lee; Yali Dou; et al.Yong ChenShu QuanUrsula Jakob Developmental ROS individualizes organismal stress resistance and lifespan. Nature 2019, 576, 301-305, 10.1038/s41586-019-1814-y.