Your browser does not fully support modern features. Please upgrade for a smoother experience.
Please note this is a comparison between Version 1 by Emil Alexov and Version 3 by Conner Chen.
Intravesicular pH plays a crucial role in melanosome maturation and function. Melanosomal pH changes during maturation from very acidic in the early stages to neutral in late stages. Neutral pH is critical for providing optimal conditions for the rate-limiting, pH-sensitive melanin-synthesizing enzyme tyrosinase (TYR). This dramatic change in pH is thought to result from the activity of several proteins that control melanosomal pH.
- pH dependence
- proton transport
- pH regulation
- stability
1. Introduction
The pH of a solution is an important characteristic for many biological processes. On a molecular level, pH controls macromolecular stability and, at extreme pH (acidic or basic extremes), macromolecules unfold. Typically, for every macromolecule, there is a particular pH at which the macromolecule is the most stable and activity is maximum, termed the pH optimum [1][2][1,2]. Macromolecular interactions are also pH-dependent [3][4][5][3,4,5], and there is typically a pH optimum at which the binding affinity is maximum [4]. Within a cell, subcellular compartments have different pH, reflecting their function, from low pH in lysosomes to high pH in peroxisomes. Thus, macromolecules tend to have a pH optimum that is ideal for the pH of the subcellular compartment where they reside [3]. Increasing the scale of this idea, pH plays a crucial role for body organ function and varies from very acidic in the stomach to neutral in the blood. All above examples indicate that the regulation and maintenance of pH is essential for many biological phenomena.
pH is maintained in a given cellular compartment by channels and/or pumps either by directly trafficking H+ or by indirectly affecting the local H+ concentration. These channels and/or pumps can be termed positive (increase pH) or negative (decrease pH) regulators [6][7][6,7]. Reaching and maintaining the desired pH depends on the balance of H+ flux controlled by these regulators, including passive transport across the membrane (Figure 1). One would expect that the positive regulators are most active at acidic pH and show almost no activity at basic pH since their role is to increase pH. The converse would be expected for negative regulators, whereby activity increases as the pH rises. At a particular pH, the inward and outward flux of H+ ions induced by positive and negative regulators becomes equal and the pH setpoint is established (Figure 1).
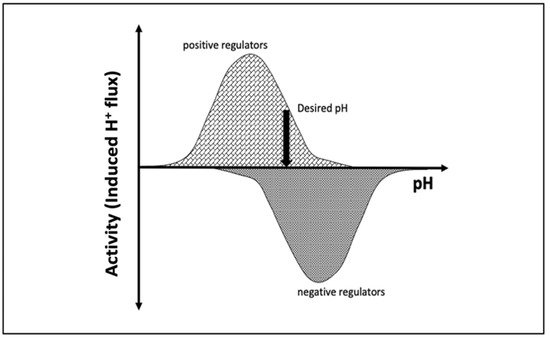
Figure 1. Schematic representation of the induced H+ flux of positive (increase pH) and negative (decrease pH) regulators. The vertical arrow indicates the desired pH, at which the total induced H+ flux is zero.
Melanocytes are a specialized cell type that resides in the skin, eyes, brain, ears, heart, lungs, and adipose tissue [8]. One of the primary functions of melanocytes is the production of melanin, a polymer of tyrosine derivatives that has important chemical properties in a wide range of tissues [9]. Melanin is synthesized in a specialized organelle called the melanosome. The pH of this organelle varies during the development of the organelle (a multistage process called maturation) and contributes to common pigmentation variation in human skin, hair, and eye color. Biallelic rare variants in proteins critical for the production of melanin (e.g., TYR) or in pH regulation of the melanosome (e.g., OCA2 and SLC45A2) lead to a significant reduction in melanin pigmentation in the skin, eyes, and hair and give rise to oculocutaneous albinism (OCA) (OCA1, OCA2, and OCA4, respectively). Melanin synthesis is critical for the protection of the skin and eyes from ultraviolet radiation; a reduction in melanin synthesis increases the risk of skin cancers. Furthermore, a dramatic reduction in melanin production in the eye is also associated with foveal hypoplasia, reduced visual acuity, and photophobia among individuals with OCA [10]. Taken together, the link between altered melanin pigment production and disease is well documented; however, it remains poorly understood how the pH of this organelle affects function of proteins critical for the maintenance of organelle pH [11].
Melanosomes originate from the endosome (Figure 2); thus, early melanosomes have a low pH (~3–4), whereas, during maturation, the pH reaches a near-neutral pH of about 7. The near-neutral pH of the mature melanosome is thought to provide a favorable environment for tyrosinase (TYR), the rate-limiting melanin-synthesizing enzyme [12][13][14][12,13,14]. The change in pH during melanosome maturation is thought to be controlled by several membrane proteins [7] (e.g., OCA2, SLC45A2, and TPC2/TPCN2) (Figure 2). OCA2 and SLC45A2 are presumed to be positive pH regulators, while TPC2 is considered to be a negative pH regulator. Considering the proposed role of positive and negative pH regulators (Figure 1), we anticipated that these proteins have different pH profiles of stability and activity. Whereas we highlight these proteins because of their association with pigment disease and published studies, it should be mentioned that other melanosome proteins may also be pH-sensitive but there are currently no genetic or functional data to support our computational examination. In addition, there are other melanosome proteins important for melanin synthesis (e.g., the ATP7A protein, which is altered in individuals with Menkes disease, and functions to supply Cu2+ to the melanosome for TYR catalytic activity) that may exhibit pH-dependent stability and activity. We hypothesized that the ATP7A protein would have a similar pH dependence to TYR [15][16][15,16].
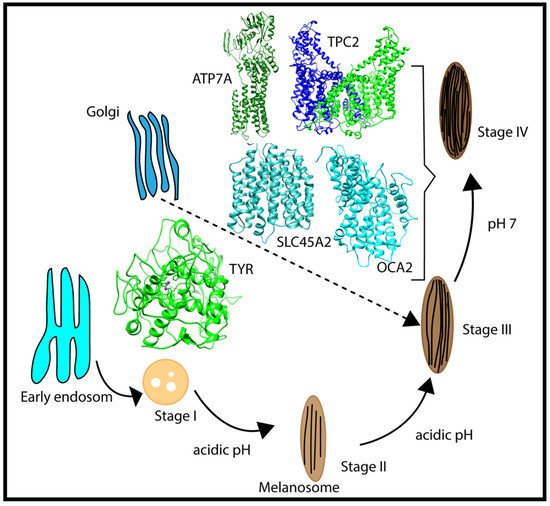
Figure 2. Schematic representation of the multistage processes of melanosome formation and proteins participating in pH regulation and melanin synthesis. The characteristic pH for each melanosome stage is also indicated in the figure.
We anticipated that OCA2 and SLC45A2 would have maximal activity at acidic pH, whereas TPC2 would have maximal activity at basic pH. OCA2 plays a major role in eye color variation [17] and regulates melanosomal pH and maturation [17][18][19][17,18,19]. It may also be involved in small-molecule transport for the biosynthesis of melanin [20][21][20,21]. SLC45A2 also participates in the transport of substances required for melanin biosynthesis [19][22][23][19,22,23]. TPC2 affects pigmentation by regulating melanosome pH and size by mediating Ca2+ release from the organelle [24][25][24,25].
2. pH Dependence of Folding Free Energy on Wild-Type Proteins
For each wild-type protein, the magnitude of the “constant” in Equation (1) is unknown, because there are no experimental data of the folding free energy (the difference of the free energy of folded and unfolded states) [26][27][28][26,27,28] at a given pH for any of the proteins modeled in this work. Accordingly, it was set to zero at the beginning of the simulated pH interval, pH = 4.0. Here, we present the calculated pH dependence of the folding free energy using an energy-minimized structure (Figure 3), and we averaged results over 20 snapshots taken from MD simulations (Figure S1). We do not focus heavily on the results obtained with MD snapshots because DelPhiPKa was developed to calculate the pKa of ionizable groups using static structures. However, we probe the sensitivity of the results using MD snapshots to investigate the role of plausible conformational changes on the pH dependence of stability. We saw no significant differences in the results obtained with the energy-minimized structure and the averaged results over 20 snapshots, suggesting that there are no structural changes contributing to the stability pH dependence.
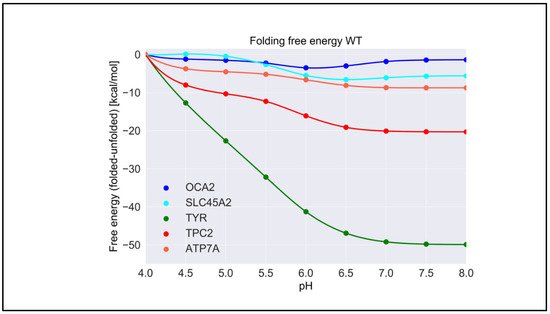

Figure 3. The pH dependence of the folding free energy of wild-type proteins from minimized structures within pH range 4–8.
TYR had the highest pH optimum of stability, about 8.0 or higher (Figure 3). In contrast, the OCA2 protein (predicted to be a positive regulator of pH) had the lowest pH optimum of stability, about 5.0–6.0. The other predicted positive regulator, the SLC45A2 protein, also had a pH optimum lower that neutral pH, i.e., 6.5. The presumed negative regulator TPC2 and the ATP7A protein which supplies copper to TYR both had pH optima close to neutral pH. Thus, there was a distinctive predicted difference in pH stability of OCA2, SLC45A2, TYR, ATP7A, and TPC2. Furthermore, our modeling confirmed the experimental observations that TYR is most active at neutral pH with reduced activity at acidic pH [14].
The pH dependence of the folding free energy is derived from the difference in pKa of ionizable groups in the folded and unfolded state. Thus, if the pKa in folded and unfolded states is the same, there would be no pH dependence. Furthermore, if the pKa is only different when outside the pH region of interest, the pH dependence of the folding free energy would still be affected but may not be physiologically relevant. It is not expected that the pKa of titratable groups in the unfolded state would be perturbed from standard pKa values [29]; thus, most of the pH dependence of the folding free energy should originate from a perturbed pKa in the folded state. However, for completeness, in Table S1, we provide the calculated pKa for both states, folded and unfolded. Indeed, one can see that, for “positive” regulators, most of perturbed pKa occurrences are for acidic groups, thus resulting in pH dependence at low pH. In contrast, most of perturbed pKa occurrences for TYR, ATP7, and the “negative” regulator TCP2 are of His residues, resulting in pH dependence at neutral pH.
3. Effect of Pathogenic Variants on Protein Stability
Table 1 shows the average change in folding free energy due to variants according to predictions made using the methods described above. The low standard deviations reported reflect the consistency of results obtained with different tools. Most of the variants were predicted to destabilize the proteins by a modest amount. However, some variants, such as A481T and N489D in OCA2, as well as C1002F and I1264V in ATP7A, were predicted to significantly affect protein stability. In the case of the OCA2 A481T and N489D variants, both of which have been observed among individuals with albinism, the predicted large change in folding free energy can be attributed to the change in the physicochemical properties of the wild-type residues: A→T and N→D. A→T represents a hydrophobic to polar residue change, while N→D represents a polar to charged residue change. In contrast, C1002F and I1264V variants in ATP7A are conservative but were also predicted to result in a large change in folding energy. In this case, the change in folding energy is thought to be caused by the distortion of the residue packing caused by the different geometries of the side-chains [30][30]. The structures of the proteins with variant sites mapped are provided in Figure S13.
Table 1.
Change in folding free energy due to variants.
| Change in Folding Free Energy (ΔΔG) Due to Variants(kcal/mol) | |||
|---|---|---|---|
| Protein | Variant | Avg ƊΔG | SD |
| TYR | R402Q | −0.5 | 0.5 |
| S192 YDouble MT * |
−0.27 −0.77 |
0.78 1.09 |
|
| OCA2 | A481T | −1.01 | 0.52 |
| H615L | 0.17 | 0.39 | |
| N489D | −1.05 | 1.08 | |
| P743L | −0.9 | 0.45 | |
| R419Q | −0.54 | 0.33 | |
| V443I | −0.54 | 0.48 | |
| SLC45A2 | G198V | −0.51 | 0.25 |
| L374F | −0.84 | 0.47 | |
| TPC2 | K376R | −0.49 | 0.3 |
| M484L | −0.86 | 0.33 | |
| M546I | −0.1 | 0.67 | |
| V219I | −0.11 | 0.32 | |
| ATP7A | C1002F | −1.2 | 0.74 |
| G666R | −0.21 | 0.7 | |
| D1044E | −0.8 | 0.53 | |
| I1264V | −1.1 | 0.74 | |
| K742R | 0.01 | 0.35 | |
| M1311V | −0.79 | 0.35 | |
| R844C | −0.48 | 0.39 | |
| S653Y | −0.45 | 0.54 | |
Note: Positive and negative values of ΔΔG represent stabilization and destabilization due to the variant, respectively. The asterisk indicates a double mutant (R402Q and S192Y) for TYR, where ΔΔG was calculated by taking the sum of individual changes.
Overall, the predicted changes in the folding free energy were not extremely large; however, since we do not know the absolute folding free energy of the proteins and how the change in protein stability affects the activity, it is impossible to definitively know how these moderate changes affect protein activity. However, we can reasonably assume that protein activity will decrease when folding free energy changes, even when the variants appear to make the protein more stable (e.g., H615L in OCA2 protein), because, in most cases, any significant deviation of wild-type properties is deleterious for protein function [31][32][31,32].
4. pH Dependence of Folding Free Energy on Genetic Variation
We predicted the effect of nonsynonymous variants on the pH dependence of protein stability by comparing the wild-type and corresponding variant proteins using both free energy-minimized structures (Figure 4) and snapshots generated via MD simulations (Figure S3). One can see that there was no significant difference in the results obtained with different protocols. Aus menthorsioned in Section 4, we considered that the “constant” in Equation (1) was the predicted folding free energy change caused by the variants (Table 1). The most drastic effects were found for OCA2, whereas variants in other proteins had moderate effects on the pH dependence of folding free energy. In the case of OCA2, most of the variants (except one, H615L) were predicted to alter the pH dependence of stability, suggesting that the variant proteins would be less stable at neutral pH.
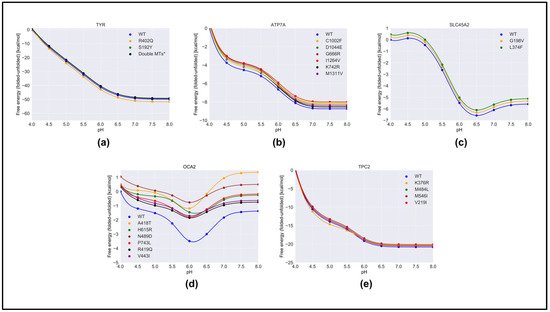

Figure 4. The pH dependence of the folding free energy of wild-type proteins and their mutants from minimized structures within pH range 4–8. (a)TYR; (b) ATP7A; (c) SLC45A2; (d) OCA2 & (e) TPC2.
Furthermore, many variants in OCA2 (R419Q, N489D, and V443I) resulted in a shift in pH optimum to lower pH. This would result in a shift in maximal activity of OCA2 toward the lower pH range and could result in a shift in the balance between positive and negative regulators such that the resulting pH setpoint would be lower than the wild-type melanosome. Lower pH in the melanosome would result in reduced TYR activity, which is found in patients with OCA2.
The above observations focused on the shape of the pH dependence curve of folding free energy without considering the magnitude of the change. It should be mentioned that the changes in the folding free energy of OCA2 were within several kcal/mol, while the changes in the pH dependence of folding free energy caused by variants in other proteins were sometimes larger (Figure 4). Despite this, the predicted changes in protein stability would likely affect protein activity and alter melanosome pH.
The reasons why variants in OCA2 had significant effects on the pH dependence of folding free energy can be found in Table S1. Our study focused on the pH interval 4.0 to 8.0, and the pH dependence was predicted to be due to titratable groups over this pH range that have different pKa values in the folded versus unfolded state. Such titratable groups are Asp, Glu, and His. One can see that, in the case of OCA2, variants resulted in a perturbed pKa of Glu and Asp, while having almost no effect on the pKa of His. This is the reason why the pH dependence of the folding free energy of OCA2 was mostly affected over acidic pH.
