Actinobacteria are among the secondary metabolites producers and hold high pharmacological and commercial interest. It has great capability to produce secondary metabolites such as immunomodulators, antibiotics, anti-cancer drugs, growth factors, anthelminthic enzymes and herbicides.describes the historical isolation of bioactive compounds from Actinobacteria from the first isolation by Selman Waksman.
- microbial ecology
- aquatic and marine environments
- drug-resistant pathogens
- natural products
- VOSviewer software
1. Introduction
Global demand for new chemotherapeutic compounds and antibiotics with high bioactivity and low toxicity has increased recently due to the emergence of life-threatening microorganisms and multidrug resistance agents among viruses, bacteria and fungi [1]. Additionally, the detection of secondary metabolites molecules with unique modes of action established various therapeutic agents’ strategies for treating many illnesses [2]. To be more specific, endophytic Actinobacteria are microorganisms that represent a new production source of a large number of secondary metabolites, including alkaloids, beta-lactams, sulfonamides, aminoglycosides, glycopeptides, siderophores, quorum-sensing molecules, immunosuppressants, polyene macrolides, saccharides, pyrazoloisoquinolinones, butenolides, nucleosides and degradative enzymes [3]. In fact, it has been reported that more than 10,000 various bioactive compounds have been discovered from Actinobacteria [4].
Endophytic microbes refer to a group of microorganisms, mostly fungi and bacteria, that exist in the host plant’s intracellular space. It usually causes no obvious harmful effect or symptoms of the disease and could produce various associations such as trophobiotic communalistic, mutualistic and symbiotic co-existence [5]. Endophytes in woody plant hosts could exist within host tissues and protect host plants against herbivores and other pathogenic microorganisms [6]. ActinobacteriaActinobacteria are Gram-positive bacteria with high guanine and cytosine (G + C) content in their genomes, and they are classified into 6 classes, 79 families of 46 orders and 10 fresh families of 16 new orders based on phylogeny using 16S rRNA sequences. The Actinobacterial classes consist of Thermoleophilia, Rubrobacteria Nitriliruptoria, Coriobacteria, Actinomycetia and Acidomicrobiia Salam et. al. [7]. Actinobacteria have ubiquitous characteristics. They are present in diverse ecosystems on the earth such as endophytically with plants and in terrestrial and aquatic environments. An abundance of Actinobacteria species have been recorded in ordinary, extraordinary and extreme environments with high or low temperatures, high radiation, acidic/alkaline pH, salinity, low levels of available moisture and nutrients [8].
The genus Streptomyces is a Gram-positive bacteria. It is the largest genus of the phylum Actinobacteria, which has complex growth and can produce various secondary metabolites [8]. In addition, there are more than 800 Streptomyces species that have been found to date (see http://www.bacterio.net/ Streptomyces .html (accessed on 20 August 2020) [9]. Streptomyces is the major microbial genus of the most antibiotic-producing bacteria in the microbial world discovered so far, where streptomycin, gentamycin, rifamycin, chloramphenicol and erythromycin are produced by Streptomyces [10].
Actinobacteria have a large number of secondary metabolite biosynthetic gene clusters. Biosynthetic gene clusters (BGCs) are known as genes comprising locally clustered groups encoding a secondary metabolite biosynthetic pathway. In addition, BGCs contain genes encoding all enzymes required to produce secondary metabolites and pathway-specific regulatory genes. The Actinobacteria have diverse physiology and metabolic flexibility with high potential to produce novel bioactive compounds and enzyme production [11].
2. Mechanism of Bioactive Compounds from Actinobacteria against Drug-Resistant Pathogens
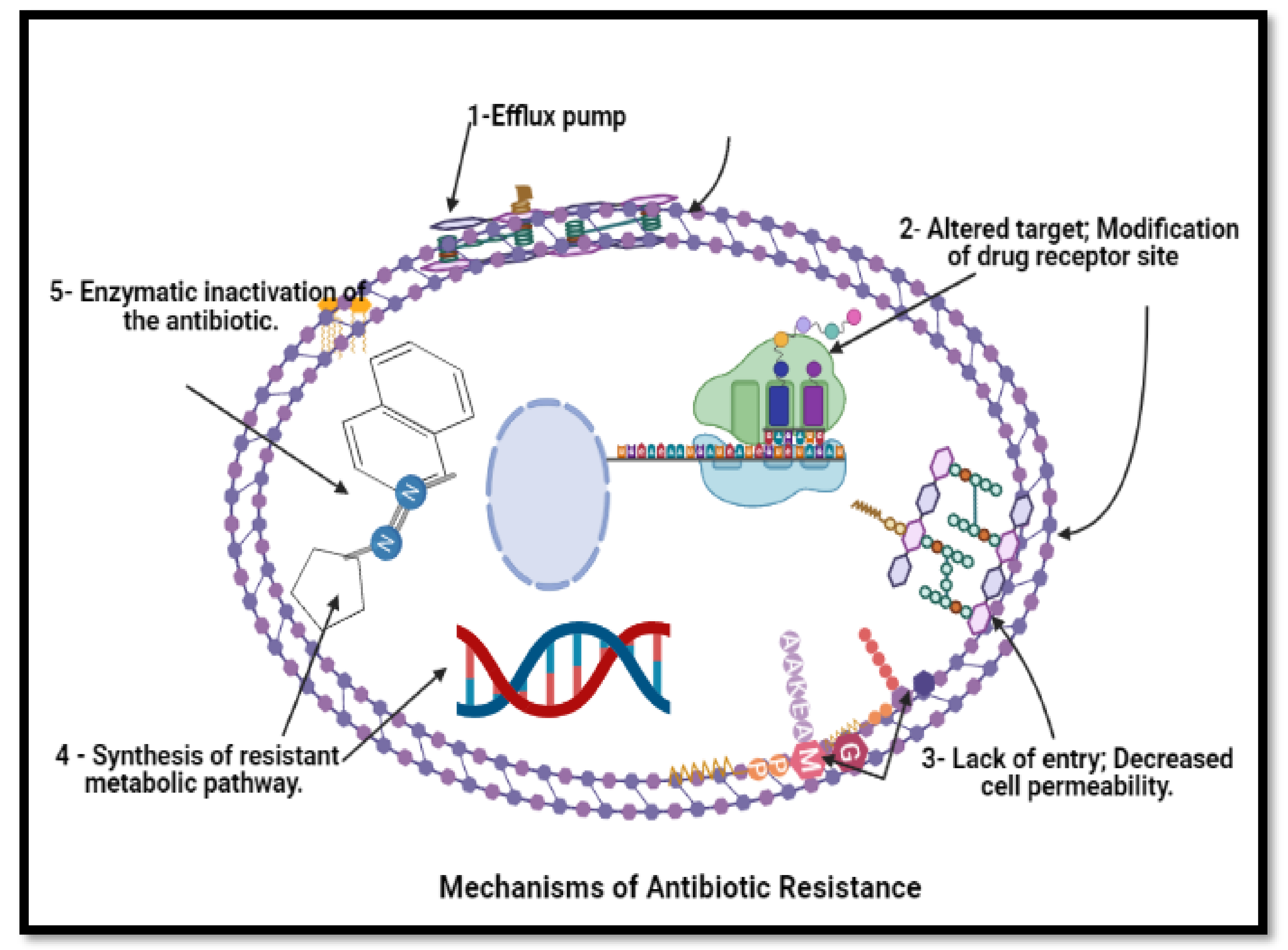 Figure 16. Mechanism of anti-bacterial resistance to avoid killing by antimicrobial molecules. N.B.: BioRender was used to draw these scientific figures.
Figure 16. Mechanism of anti-bacterial resistance to avoid killing by antimicrobial molecules. N.B.: BioRender was used to draw these scientific figures.3. Conclusions and Future Prospects
References
- Carvalho, I.T.; Santos, L. Antibiotics in the aquatic environments: A review of the European scenario. Environ. Int. 2016, 94, 736–757.
- Ribeiro, I.; Girão, M.; Alexandrino, D.; Ribeiro, T.; Santos, C.; Pereira, F.; Mucha, A.; Urbatzka, R.; Leão, P.; Carvalho, M. Diversity and Bioactive Potential of Actinobacteria Isolated from a Coastal Marine Sediment in Northern Portugal. Microorganisms 2020, 8, 1691.
- Qi, D.; Zou, L.; Zhou, D.; Chen, Y.; Gao, Z.; Feng, R.; Zhang, M.; Li, K.; Xie, J.; Wang, W. Taxonomy and Broad-Spectrum Antifungal Activity of Streptomyces sp. SCA3-4 Isolated From Rhizosphere Soil of Opuntia stricta. Front. Microbiol. 2019, 10, 1390.
- Girão, M.; Ribeiro, I.; Ribeiro, T.; Azevedo, I.C.; Pereira, F.; Urbatzka, R.; Leão, P.; Carvalho, M.F. Actinobacteria Isolated From Laminaria ochroleuca: A Source of New Bioactive Compounds. Front. Microbiol. 2019, 10, 683.
- Kandel, S.L.; Joubert, P.M.; Doty, S.L. Bacterial Endophyte Colonization and Distribution within Plants. Microorganisms 2017, 5, 77.
- Frank, A.C.; Guzmán, J.P.S.; Shay, J.E. Transmission of Bacterial Endophytes. Microorganisms 2017, 5, 70.
- Salam, N.; Jiao, J.-Y.; Zhang, X.-T.; Li, W.-J. Update on the classification of higher ranks in the phylum Actinobacteria. Int. J. Syst. Evol. Microbiol. 2020, 70, 1331–1355.
- Passari, A.K.; Chandra, P.; Leo, V.V.; Mishra, V.K.; Kumar, B.; Singh, B.P. Production of potent antimicrobial compounds from Streptomyces cyaneofuscatus associated with fresh water sediment. Front Microbiol. 2017, 8, 68.
- Li, L.-Y.; Yang, Z.-W.; Asem, M.D.; Fang, B.-Z.; Salam, N.; Alkhalifah, D.H.M.; Hozzein, W.N.; Nie, G.-X.; Li, W.-J. Streptomyces desertarenae sp. nov., a novel actinobacterium isolated from a desert sample. Antonie Leeuwenhoek 2019, 112, 367–374.
- Bérdy, J. Bioactive Microbial Metabolites. J. Antibiot. 2005, 58, 1–26.
- Shivlata, L.; Satyanarayana, T. Thermophilic and alkaliphilic Actinobacteria: Biology and potential applications. Front. Microbiol. 2015, 6, 1014.
- Yamaguchi, M.; Goto, K.; Hirose, Y.; Yamaguchi, Y.; Sumitomo, T.; Nakata, M.; Nakano, K.; Kawabata, S. Identification of evolutionarily conserved virulence factor by selective pressure analysis of Streptococcus pneumoniae. Commun. Biol. 2019, 2, 96.
- Lee, W.; Do, T.; Zhang, G.; Kahne, D.; Meredith, T.C.; Walker, S. Antibiotic Combinations That Enable One-Step, Targeted Mutagenesis of Chromosomal Genes. ACS Infect. Dis. 2018, 4, 1007–1018.
- Schwarz, S.; Loeffler, A.; Kadlec, K. Bacterial resistance to antimicrobial agents and its impact on veterinary and human medicine. Adv. Vet. Dermatol. 2017, 8, 95–100.
- Touchon, M.; Sousa, J.A.M.D.; Rocha, E.P. Embracing the enemy: The diversification of microbial gene repertoires by phage-mediated horizontal gene transfer. Curr. Opin. Microbiol. 2017, 38, 66–73.
- Kapoor, G.; Saigal, S.; Elongavan, A. Action and resistance mechanisms of antibiotics: A guide for clinicians. J. Anaesthesiol. Clin. Pharmacol. 2017, 33, 300–305.
- Uma Maheswari, R. Phenotypic and Genotypic Characterisation of Vancomycin Resistant Staphylococcus aureus among MRSA Isolates in a Tertiary Care Hospital. Master’s Thesis, Tirunelveli Medical College, Tirunelvel, India, 2019. Available online: http://repository-tnmgrmu.ac.in/id/eprint/11129 (accessed on 23 August 2019).
