Prostate cancer biomarkers can be measured in urine, blood or tissue. A variety of tests that analyse patients' biomarkers have been developed to improve diagnosis, prognosis and to help stratify individual's risk of prostate cancers. Liquid biopsy biomarkers are easy-to-use and non-invasive. They guide the decision-making process, determine whether the patient requires treatment or can be monitored under active surveillance, and help choose the best treatment option.
- cancer biomarkers
- prostate cancer
- liquid biopsy
- prognosis
- diagnosis
- early detection
- prostate biopsy
- non-invasive
- urology
- urine biomarkers
1. Introduction: Prostate Cancer Diagnosis
The traditional diagnosis of PCa is based on the assessment of serum prostate-specific antigen (PSA) levels, digital rectal examination (DRE), followed by biopsy under the guidance of transrectal ultrasonography (TRUS). Serum PSA levels are commonly used for detection, risk stratification, and monitoring of PCa [1]. In screening programs, high PSA levels, despite a normal DRE, lead to the diagnosis of PCa in more than 60% of asymptomatic patients. Unfortunately, it results in a high number of unnecessary biopsies and the detection of asymptomatic cancers with low clinical risk [2]
Currently, the gold-standard test to confirm cancer is the histopathological result of a prostate biopsy. Unfortunately, this invasive procedure is painful, expensive, and may pose a risk of complications (e.g., infection or sepsis). Furthermore, the procedure is prone to significant sampling error. It is therefore essential to avoid unnecessary biopsies [3][4].
This highlights the need to develop more precise methods to identify clinically relevant PCa, such as liquid biopsy-derived biomarkers.
Liquid biopsy is a minimally invasive procedure that analyses even small targets, which allows its usefulness in screening, diagnosis, prognosis, follow-up and therapeutic management [5]. Prostate Cancer biomarkers can be detected in blood, urine and tissue (Figure1.)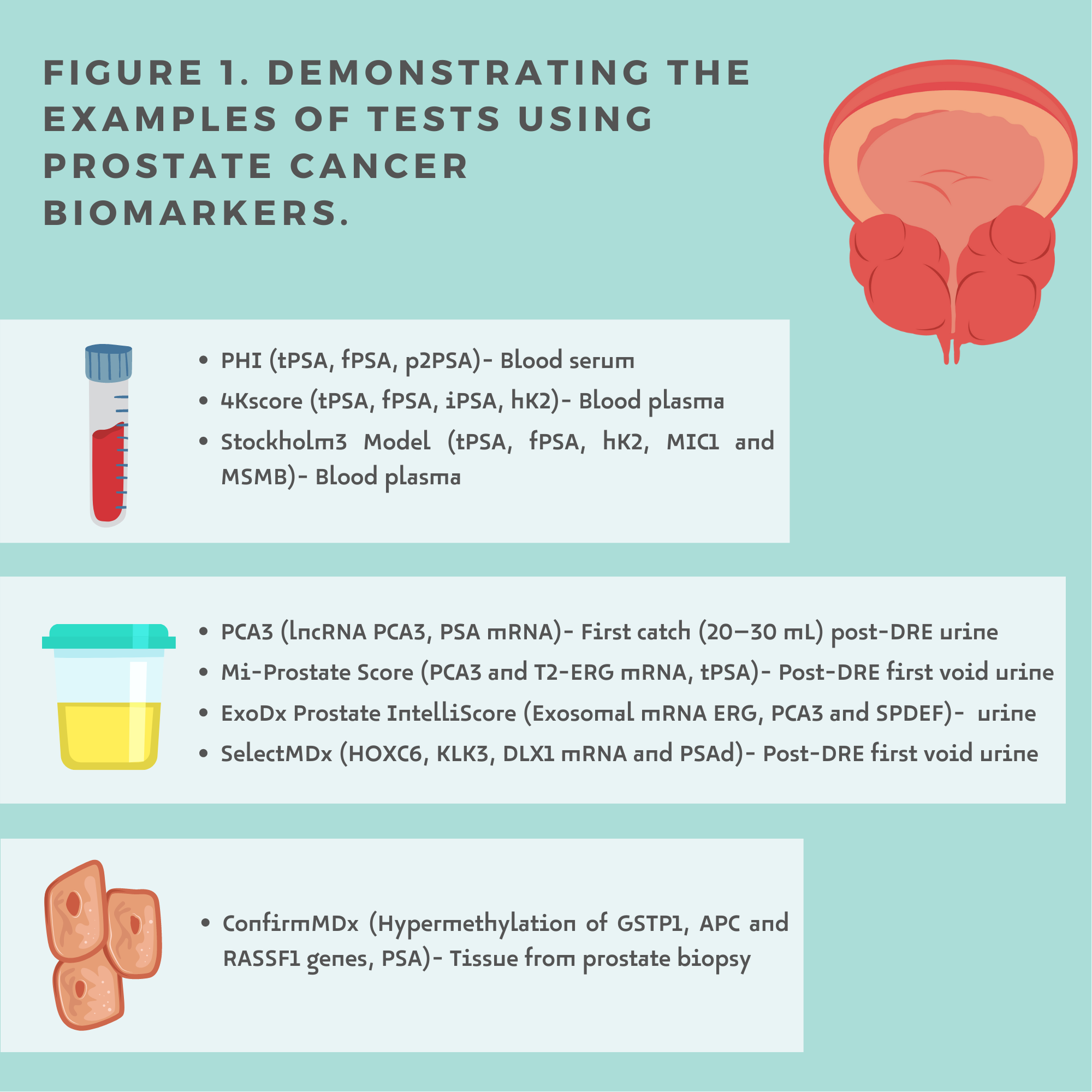
Liquid biopsy biomarkers are proving to be a promising new diagnostic and prognostic approach to help optimize the pre-biopsy decision and stratify whether the patient requires treatment or can be monitored under active surveillance.
There are the following types of biomarkers (Figure 2.):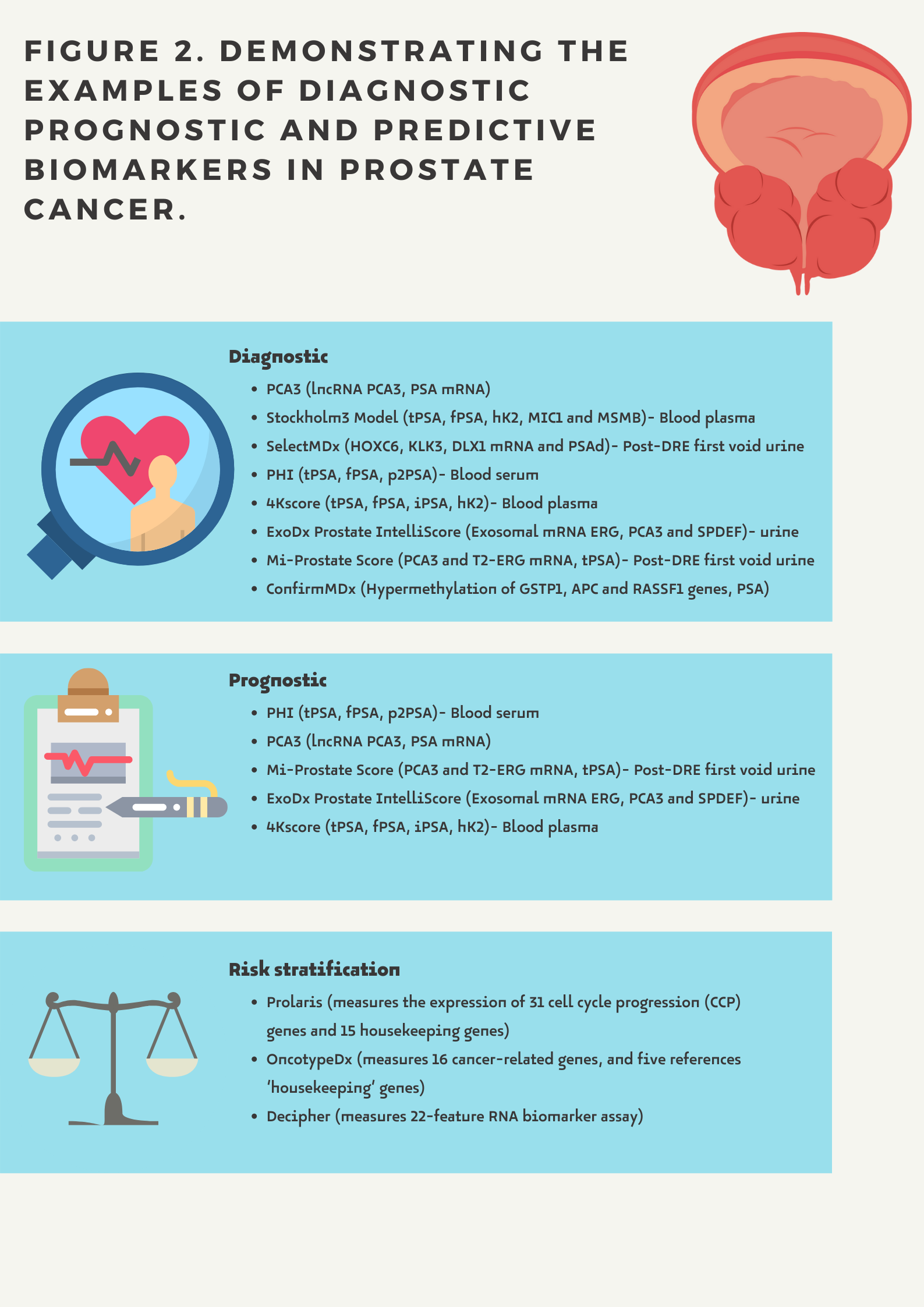
- Diagnostic biomarkers provide information about the potential risk of prostate cancer.
- Prognostic biomarkers show the possible course of the disease in a given patient regardless of whether or not a treatment is administered. A prognostic biomarker indicates an increased (or decreased) likelihood of a future clinical event, disease recurrence or progression for an individual patient
- Predictive biomarkers related to the assessment of a patient's response to a given treatment and indicating the likelihood of a particular outcome depending on whether a therapeutic intervention is undertaken (e.g. hormonal status)
2. Urine Biomarkers
Urine is obtained non-invasively and contains fluid excreted from the prostate gland, including products from prostate cancer cells [6]. For many urinary biomarkers, performing a DRE is crucial as it increases fluid excretion from the prostate. To date, four prostate cancer tests are available with proven clinical utility.
The Progensa PCA3 test (Hologic Inc., Marlborough, MA, USA) is a test that measures PSA messenger RNA (mRNA) and PCA3 mRNA detectable in the first catch urine sample after DRE.
It is the first urine biomarker test approved in 2006 by the European Union, Canada [7] and in 2012 by the FDA. The FDA recommends its use in men ≥ 50 years old to support repeat biopsy decision-making in whom one or more previous prostate biopsies have been negative and for whom repeat biopsy is recommended based on current standards of care [8]. However, some clinical studies [9][10] report the benefits of using the test as early as the first biopsy.
Numerous studies [10][11] have shown that the diagnostic value of the test increases when adding other predictors (i.e., age, PSA value, DRE result or prostate volume). Therefore, the producer recommends its use in combination with standard diagnostic parameters [12].
MyProstateScore (MPS, LynxDx, Ann Arbor, MI, USA (previously known as MiPS—Michigan Prostate Score)) is an algorithm that measures mRNA, PCA3 and TMPRSS-ERG (abnormal fusion of TMPRSS2 and ERG (T2-ERG)) expression in urine from the first collection after DRE and serum PSA.
More than 50% of patients with PCa have an ERG gene fusion with TMPRSS2 [13]. This translocation is associated with poor patient prognosis—an increased risk of recurrence and mortality from PCa.
The test is indicated for men with suspicious PSA levels who are considered for initial or repeat biopsy. The test result validates the need for biopsy and predicts the risk of high-grade prostate cancer (GS > 7) in a diagnostic needle biopsy [14][15][16]. Values range from 0 to 100, and the higher the score, the greater the risk of aggressive cancer.
The ExoDx Prostate (IntelliScore) (EPI, Exosome Diagnostics, Waltham, MA, USA) assesses the exosomal RNA expression of three genes (ERG, PCA3 and SPDEF) involved in the initiation and progression of PCa. ExoDx prostate is a test performed from a urine sample that does not require prior DRE testing. Exosomal RNA is derived from exosomes, small membrane vesicles secreted by several cell types, including immune and cancer cells [17]. The high potential of exosomes as biomarkers is due to their structure—a lipid bilayer protects the contents from degradation by proteases.
The test scores range from 1 to 100, and a cut-off point of 15.6 indicates men at increased risk of HG PCa (≥GG2) at subsequent biopsy, making the test helpful in validating the need for biopsy in men at risk. The test is recommended for men aged ≥50 years who are in the PSA "grey zone" (2–10 ng/mL) to distinguish a benign (HG1; when the test value < 15.6) from high-grade PCa (HG2 ≥ 15.6) [18]
The study [19] of PCa biomarkers with a blinded control arm showed that ExoDx helped avoid unnecessary biopsies when the test was negative and increased the detection of HG PCa by 30% compared with a control arm without ExoDx (SOC alone). Compared to SOC, the test missed 49% fewer HG PCa. The study showed that ExoDx improved patient stratification and influenced the decisions made by (68%) urologists about biopsy (with rising PSA being the main reason for not following ExoDx results).
SelectMDx (MDxHealth, Inc., Irvine, CA, USA) is a urine-based test after DRE that measures three biomarkers: DLX1 (progression gene), HOXC6 (cell proliferation gene), KLK3 (reference gene) and clinical risk factors (age, DRE, PSA and prostate volume which can be calculated from the TRUS measurements substituted into the formula: height × width × length × 0.523). HOXC6 and DLX1 mRNA levels are assessed to estimate the risk of PCa on biopsy and the presence of high-risk cancer.
Men with elevated PSA levels in the "grey zone" (4–10 ng/mL) and/or an abnormal DRE result are subjects for whom an initial biopsy is considered. The result determines whether the patient is at high or low risk of PCa. It supports clinical decision-making and stratifies patients into those who may benefit from biopsy and early cancer detection and others for whom it is better to avoid this invasive procedure and continue with routine screening or active surveillance.
3. Serum Biomarkers
Serum biomarkers, determined from blood samples, are produced by healthy and abnormal cells. PSA is undoubtedly the most widely studied cancer biomarker, but its clinical utility due to low specificity and specificity raises the need to find a test with better diagnostic values. Three tests that may have a positive impact on clinical practice are described below.
The Prostate Health Index (PHI; Beckman Coulter Inc., Brea, CA, USA), determined from a serum test, includes total PSA (tPSA), free, non-protein-bound PSA (fPSA) and (–2)proPSA (the fPSA isoform resulting from incomplete processing of the PSA precursor).
Determination of PHI values is indicated in men with PSA levels in the "grey zone" (4–10 ng /mL) and an unsuspected digital rectal examination (DRE) result [20], at age ≥50 years. The PHI score is calculated from the formula: ((–2)proPSA/fPSA) × √PSA.
High PHI values indicate an increased likelihood of detecting prostate cancer, so when a biopsy (initial or repeat) is recommended, consideration should be given to using this less invasive method.
The PHI score has a better diagnostic value than considering each of the indices (tPSA, fPSA) separately [21][22] improves the detection of PCa [23] improves clinical decision-making and predicts PCa aggressiveness [21][24][25].
The 4Kscore (Opko Health Inc., Miami, FL, USA) is a test developed to identify HG-PCa in patients with a suspicious DRE or elevated serum PSA. total PSA (tPSA), free PSA (fPSA), intact PSA (iPSA) and serum levels of human kallikrein 2 (hK2). It then compares the values obtained in the algorithm with the patient's age, DRE and results of previous prostate biopsies. However, it can be performed by any man over 35 years of age who wants to assess his personalized risk of disease in the future.
The 4K test, although not designed to assess the predicted course of already diagnosed prostate cancer, has also been used in patients with csPCa to identify candidates for more intensive therapy. It has also been used to improve treatment selection and thus increase the chance of cure in patients suspected of having an underestimated malignancy. The 4Kscore provides an estimate of a patient's risk of developing distant metastases within ten years.
Stockholm 3 Model (S3M) combines serum biomarkers (total PSA, free PSA, free/total PSA ratio, hK2, MIC1 and MSMB with genetic markers (254 single nucleotide polymorphisms [SNPs] and an unclassified variable for SNP HOXB13). The test also considers clinical data (age, previous prostate biopsy—family history, use of 5-alpha reductase inhibitors) and prostate examination (DRE, prostate volume). The S3M available in Sweden, Norway, Denmark, and Finland is in clinical use to predict the risk of aggressive prostate cancer and assess the need for biopsy. The S3M research team at Karolinska Institute is currently working with two major laboratories in Europe and laboratories in the US and Canada to introduce the test in additional countries around the world.
Studies [26][27][28][29][30] prove that S3M reduces overdiagnosis and the number of prostate biopsies while maintaining sensitivity for clinically significant prostate cancer.
Long-term follow-up of PSA replacement (as part of the standard prostate cancer diagnostic procedure) with Stockholm3 in prostate cancer detection in primary care in the Stavanger region of Norway showed that the implementation was beneficial. Compared with PSA, S3M reduced the proportion of clinically insignificant PCa (from 58% to 35%) and the number of biopsies performed (from 29.0% to 20.8%). This management may also lead to a reduction in healthcare costs. It has been estimated that direct healthcare costs decreased by 23–28% per male studied [31].
S3M is not suitable for men who have previously been diagnosed with or treated for prostate cancer or under follow-up after prostate cancer. It has no proven value for men diagnosed with prostate cancer or who have undergone a biopsy or other examination by a urologist within the last six months. The S3M was shown to be superior to prostate-specific antigen (PSA) as a screening tool for prostate cancer in all men aged 50–70 years. The S3M may reduce unnecessary biopsies without compromising the ability to diagnose prostate cancer with a Gleason score of at least 7.
4. Tissue Biomarkers
Tissue biomarkers analyze changes in nucleic acid expression and composition of tissue collected during needle core biopsy of the prostate. The main concept is to detect changes in the histologically normal field neighbouring prostate cancer. This helps to verify whether the patient requires an additional biopsy.
ConfirmMDx (MDxHealth)
ConfirmMDx (MDxHealth, Inc., Irvine, CA, USA) is a tissue-based epigenetic assay that uses methylation-specific PCR (MSP) to analyze three prostate-cancer-related changes in DNA methylation patterns of suppressor genes (GSTP1, APC and RASSF1) in biopsy tissue (formalin-fixed and paraffin-embedded). All these biomarkers were isolated from biopsy-positive tissue.
ConfirmMDx is a molecular test clinically validated for predicting prostate cancer risk in men who have had a traditional thick-needle prostate biopsy that did not reveal the presence of cancer cells in collected histopathological material. In many cases, prostate biopsy results are falsely negative. The biopsy specimen may not be cancerous, and the histopathological result will not reveal the presence of cancer. However, due to the "halo effect", tissue with a normal morphological appearance will show epigenetic changes, indicating the presence of cancer. Using the test, histopathological material that has already been taken—during a prostate biopsy—can be re-examined in a detailed epigenetic analysis quantifying the level of methylation of promoter regions of three genes in benign prostate tissue assessing with high accuracy the presence of cancer cells in neighbouring areas.
ConfirmMDx offers the opportunity to avoid unnecessary repeat biopsies. It allows a decision to be made on whether to include (rule-in) or exclude (rule-out) therapy. High-risk men with a previously negative biopsy may have undetected cancer after the test. Patients with a previous "false negative" biopsy result should be included for repeat biopsy and appropriate treatment.
It also allows low-risk men to be excluded from repeat biopsies, protecting the patient from unnecessary stress and possible complications and reducing healthcare costs. This test increases the negative predictive value.
Recently, molecular characterization of PCa has become increasingly important, and a wide range of biomarker-based liquid biopsy tests are commercially available to assist urologists in clinical decision-making. The prostate cancer liquid biopsy biomarkers listed above have a high NPV and therefore help prevent unnecessary biopsies.
From a clinical point of view, it is critical to identify assays for the early detection of aggressive PCa subtype when it can still be treated effectively. Recent years have led to the development of totally non-invasive tests, i.e., (ExoDx Prostate At-Home Collection) where first catch, nondigital rectal examination urine specimens appeared helpful in identifying aggressive (Gleason score 7–10) PCa in a racially diverse patient cohort. Similarly, the four-kallikrein panel showed effectiveness in identifying aggressive PCa in a multiethnic population.
Current research focuses on comparing biomarker tests with mpMRI and on the extent to which they can complement each other.
It seems that soon, molecular biomarkers, clinical and histopathological features and diagnostic imaging will have to be used in a complementary rather than a competitive manner to ensure the best possible selection of patients for mpMRI and eventual biopsy.
The content is from 10.3390/cancers13133373
References
- Lilja, H.; Ulmert, D.; Vickers, A.J. Prostate-specific antigen and prostate cancer: Prediction, detection and monitoring. Nature reviews. Cancer 2008, 8, 268–278. [Google Scholar] [PubMed]
- Graif, T.; Loeb, S.; Roehl, K.A.; Gashti, S.N.; Griffin, C.; Yu, X.; Catalona, W.J. Under Diagnosis and Over Diagnosis of Prostate Cancer. J. Urol. 2007, 178, 88–92. [Google Scholar] [CrossRef] [PubMed]
- Lundström, K.J.; Drevin, L.; Carlsson, S.; Garmo, H.; Loeb, S.; Stattin, P.; Bill-Axelson, A. Nationwide population based study of infections after transrectal ultrasound guided prostate biopsy. J. Urol. 2014, 192, 1116–1122. [Google Scholar] [CrossRef] [PubMed]
- Bruyère, F.; Malavaud, S.; Bertrand, P.; Decock, A.; Cariou, G.; Doublet, J.D.; Bernard, L.; Bugel, H.; Conquy, S.; Sotto, A.; et al. Prosbiotate: A Multicenter, Prospective Analysis of Infectious Complications after Prostate Biopsy. J. Urol. 2015, 193, 145–150. [Google Scholar] [CrossRef] [PubMed]
- Fernández-Lázaro, D.; Hernández, J.L.G.; García, A.C.; Martínez, A.C.; Mielgo-Ayuso, J. Liquid Biopsy as Novel Tool in Precision Medicine: Origins, Properties, Identification and Clinical Perspective of Cancer’s Biomarkers. Diagnostics 2020, 10, 215. [Google Scholar] [CrossRef]
- Hessels, D.; Klein Gunnewiek, J.M.T.; van Oort, I.; Karthaus, H.F.; van Leenders, G.J.; van Balken, B.; Kiemeney, L.; Witjes, J.; Schalken, J.A. DD3PCA3-based Molecular Urine Analysis for the Diagnosis of Prostate Cancer. Eur. Urol. 2003, 44, 8–16. [Google Scholar] [CrossRef]
- Pal, R.P.; Maitra, N.U.; Mellon, J.K.; Khan, M.A. Defining prostate cancer risk before prostate biopsy. Urol. Oncol. Semin. Orig. Investig. 2013, 31, 1408–1418. [Google Scholar] [CrossRef]
- Marks, L.S.; Fradet, Y.; Deras, I.L.; Blase, A.; Mathis, J.; Aubin, S.M.; Cancio, A.T.; Desaulniers, M.; Ellis, W.J.; Rittenhouse, H.; et al. PCA3 Molecular Urine Assay for Prostate Cancer in Men Undergoing Repeat Biopsy. Urology 2007, 69, 532–535. [Google Scholar] [CrossRef] [PubMed]
- . Rodríguez, S.V.M.; García-Perdomo, H.A. Diagnostic accuracy of prostate cancer antigen 3 (PCA3) prior to first prostate biopsy: A systematic review and meta-analysis. Can. Urol. Assoc. J. 2019, 14, E214–E219. [Google Scholar] [CrossRef] [PubMed]
- Wei, J.T.; Feng, Z.; Partin, A.W.; Brown, E.; Thompson, I.; Sokoll, L.; Chan, D.W.; Lotan, Y.; Kibel, A.S.; Busby, J.E.; et al. Can Urinary PCA3 Supplement PSA in the Early Detection of Prostate Cancer? J. Clin. Oncol. 2014, 32, 4066–4072. [Google Scholar] [CrossRef] [PubMed]
- Aubin, S.M.; Reid, J.; Sarno, M.J.; Blase, A.; Aussie, J.; Rittenhouse, H.; Rittmaster, R.; Andriole, G.L.; Groskopf, J. PCA3 Molecular Urine Test for Predicting Repeat Prostate Biopsy Outcome in Populations at Risk: Validation in the Placebo Arm of the Dutasteride REDUCE Trial. J. Urol. 2010, 184, 1947–1952. [Google Scholar] [CrossRef]
- Progensa PCA3 Assay. Available online: https://www.hologic.com/sites/default/files/2019-05/502083-IFU-PI_003_01.pdf (accessed on 20 December 2020).
- Tomlins, S.A.; Rhodes, D.R.; Perner, S.; Dhanasekaran, S.M.; Mehra, R.; Sun, X.-W.; Varambally, S.; Cao, X.; Tchinda, J.; Kuefer, R.; et al. Recurrent Fusion of TMPRSS2 and ETS Transcription Factor Genes in Prostate Cancer. Science 2005, 310, 644–648. [Google Scholar] [CrossRef] [PubMed]
- Leyten, G.H.J.M.; Hessels, D.; Jannink, S.A.; Smit, F.P.; de Jong, H.; Cornel, E.B.; de Reijke, T.M.; Vergunst, H.; Kil, P.; Knipscheer, B.C.; et al. Prospective Multicentre Evaluation of PCA3 and TMPRSS2-ERG Gene Fusions as Diagnostic and Prognostic Urinary Biomarkers for Prostate Cancer. Eur. Urol. 2014, 65, 534–542. [Google Scholar] [CrossRef]
- Tomlins, S.A.; Day, J.R.; Lonigro, R.J.; Hovelson, D.; Siddiqui, J.; Kunju, L.P.; Dunn, R.L.; Meyer, S.; Hodge, P.; Groskopf, J.; et al. Urine TMPRSS2:ERG Plus PCA3 for Individualized Prostate Cancer Risk Assessment. Eur. Urol. 2016, 70, 45–53. [Google Scholar] [CrossRef]
- Salami, S.S.; Schmidt, F.; Laxman, B.; Regan, M.M.; Rickman, D.S.; Scherr, D.; Bueti, G.; Siddiqui, J.; Tomlins, S.A.; Wei, J.T.; et al. Combining urinary detection of TMPRSS2:ERG and PCA3 with serum PSA to predict diagnosis of prostate cancer. Urol. Oncol. Semin. Orig. Investig. 2013, 31, 566–571. [Google Scholar] [CrossRef]
- Denzer, K.; Kleijmeer, M.J.; Heijnen, H.F.; Stoorvogel, W.; Geuze, H.J. Exosome: From internal vesicle of the multivesicular body to intercellular signaling device. J. Cell Sci. 2000, 113, 3365–3374. [Google Scholar] [CrossRef] [PubMed]
- McKiernan, J.; Donovan, M.J.; O’Neill, V.; Bentink, S.; Noerholm, M.; Belzer, S.; Skog, J.; Kattan, M.W.; Partin, A.; Andriole, G.; et al. A Novel Urine Exosome Gene Expression Assay to Predict High-grade Prostate Cancer at Initial Biopsy. JAMA Oncol. 2016, 2, 882–889. [Google Scholar] [CrossRef]
- Tutrone, R.; Donovan, M.J.; Torkler, P.; Tadigotla, V.; McLain, T.; Noerholm, M.; Skog, J.; McKiernan, J. Clinical utility of the exosome based ExoDx Prostate(IntelliScore) EPI test in men presenting for initial Biopsy with a PSA 2–10 ng/mL. Prostate Cancer Prostatic Dis. 2020, 23, 607–614. [Google Scholar] [CrossRef]
- Loeb, S.; Sanda, M.G.; Broyles, D.L.; Shin, S.S.; Bangma, C.H.; Wei, J.T.; Partin, A.; Klee, G.G.; Slawin, K.M.; Catalona, W.J.; et al. The Prostate Health Index Selectively Identifies Clinically Significant Prostate Cancer. J. Urol. 2015, 193, 1163–1169. [Google Scholar] [CrossRef]
- Catalona, W.J.; Partin, A.W.; Sanda, M.G.; Wei, J.T.; Klee, G.G.; Bangma, C.H.; Slawin, K.M.; Marks, L.S.; Loeb, S.; Broyles, D.L.; et al. A Multicenter Study of (–2)Pro-Prostate Specific Antigen Combined with Prostate Specific Antigen and Free Prostate Specific Antigen for Prostate Cancer Detection in the 2.0 to 10.0 ng/ml Prostate Specific Antigen Range. J. Urol. 2011, 185, 1650–1655. [Google Scholar] [CrossRef]
- Boegemann, M.; Stephan, C.; Cammann, H.; Vincendeau, S.; Houlgatte, A.; Jung, K.; Blanchet, J.-S.; Semjonow, A. The percentage of prostate-specific antigen (PSA) isoform (–2)proPSA and the Prostate Health Index improve the diagnostic accuracy for clinically relevant prostate cancer at initial and repeat biopsy compared with total PSA and percentage free PSA in men. BJU Int. 2015, 117, 72–79. [Google Scholar] [CrossRef]
- Stephan, C.; Kahrs, A.-M.; Cammann, H.; Lein, M.; Schrader, M.; Deger, S.; Miller, K.; Jung, K. A (–2)proPSA-based artificial neural network significantly improves differentiation between prostate cancer and benign prostatic diseases. Prostate 2009, 69, 198–207. [Google Scholar] [CrossRef]
- Jansen, F.H.; van Schaik, R.H.; Kurstjens, J.; Horninger, W.; Klocker, H.; Bektic, J.; Wildhagen, M.F.; Roobol, M.J.; Bangma, C.H.; Bartsch, G. Prostate-Specific Antigen (PSA) Isoform p2PSA in Combination with Total PSA and Free PSA Improves Diagnostic Accuracy in Prostate Cancer Detection. Eur. Urol. 2010, 57, 921–927. [Google Scholar] [CrossRef] [PubMed]
- Guazzoni, G.; Nava, L.; Lazzeri, M.; Scattoni, V.; Lughezzani, G.; Maccagnano, C.; Dorigatti, F.; Ceriotti, F.; Pontillo, M.; Bini, V.; et al. Prostate-Specific Antigen (PSA) Isoform p2PSA Significantly Improves the Prediction of Prostate Cancer at Initial Extended Prostate Biopsies in Patients with Total PSA Between 2.0 and 10 ng/ml: Results of a Prospective Study in a Clinical Setting. Eur. Urol. 2011, 60, 214–222. [Google Scholar] [CrossRef]
- Grönberg, H.; Adolfsson, J.; Aly, M.; Nordström, T.; Wiklund, P.; Brandberg, Y.; Thompson, J.; Wiklund, F.; Lindberg, J.; Clements, M.; et al. Prostate cancer screening in men aged 50–69 years (STHLM3): A prospective population-based diagnostic study. Lancet Oncol. 2015, 16, 1667–1676. [Google Scholar] [CrossRef]
- Ström, P.; Nordström, T.; Aly, M.; Egevad, L.; Grönberg, H.; Eklund, M. The Stockholm-3 Model for Prostate Cancer Detection: Algorithm Update, Biomarker Contribution, and Reflex Test Potential. Eur. Urol. 2018, 74, 204–210. [Google Scholar] [CrossRef]
- Möller, A.; Olsson, H.; Grönberg, H.; Eklund, M.; Aly, M.; Nordström, T. The Stock-holm3 blood-test predicts clinically-significant cancer on biopsy: Independent validation in a multi-center community cohort. Prostate Cancer Prostatic Dis. 2018, 22, 137–142. [Google Scholar] [CrossRef]
- Eklund, M.; Nordström, T.; Aly, M.; Adolfsson, J.; Wiklund, P.; Brandberg, Y.; Thompson, J.; Wiklund, F.; Lindberg, J.; Presti, J.C.; et al. The Stockholm-3 (STHLM3) Model can Improve Prostate Cancer Diagnostics in Men Aged 50–69 yr Compared with Current Prostate Cancer Testing. Eur. Urol. Focus 2018, 4, 707–710. [Google Scholar] [CrossRef]
- Nordström, T.; Grönberg, H.; Adolfsson, J.; Egevad, L.; Aly, M.; Eklund, M. Balancing Overdiagnosis and Early Detection of Prostate Cancer using the Stockholm-3 Model. Eur. Urol. Focus 2018, 4, 385–387. [Google Scholar] [CrossRef]
- Viste, E.; Vinje, C.A.; Lid, T.G.; Skeie, S.; Evjen-Olsen, Ø.; Nordström, T.; Thorsen, O.; Gilje, B.; Janssen, E.A.M.; Kjosavik, S.R. Effects of replacing PSA with Stockholm3 for diagnosis of clinically significant prostate cancer in a healthcare system—The Stavanger experience. Scand. J. Prim. Heal. Care 2020, 38, 315–322. [Google Scholar] [CrossRef]
