Recently, a new class of prokaryotic compartments, collectively called encapsulins or protein nanocompartments, has been discovered. The shell proteins of these structures self-organize to form icosahedral compartments with a diameter of 25–42 nm, while one or more cargo proteins with various functions can be encapsulated in the nanocompartment. Non-native cargo proteins can be loaded into nanocompartments and the surface of the shells can be further functionalized, which allows for developing targeted drug delivery systems or using encapsulins as contrast agents for magnetic resonance imaging. Since the genes encoding encapsulins can be integrated into the cell genome, encapsulins are attractive for investigation in various scientific fields, including biomedicine and nanotechnology.
- encapsulin
- nanocompartment
- cell labeling
- MRI
1. Introduction
2. Structural Organization of the Shell
Encapsulin shells are icosahedral (12 vertices, 20 faces, 30 edges) complexes formed by self-assembly of protomer proteins [2,13,24][2][13][24]. Encapsulin shell proteins are homologous to gp5-HK97 phage main capsid protein (Figure 1A), as evidenced by the structural similarity between the gp5 protomer protein and encapsulin shell protein [26]. It should be noted that the HK97 fold is widespread in nature and has been observed in all other tailed phages [27], in herpesvirus capsids [28], and in the archaevirus HSTV-1 [29]. Like gp5, the encapsulin shell proteins oligomerize to form a complete nanocompartment. As well as viral capsids, encapsulin shell proteins can self-assemble into icosahedrons of different sizes. For example, encapsulins from Pyrococcus furiosus and Myxococcus xanthus consist of 180 protomers (30‒32 nm in diameter) (Figure 1C,D), while those from Thermotoga maritima are composed of 60 protomers (24 nm in diameter) (Figure 1B) and encapsulins from Quasibacillus thermotolerans consist of 240 protomers (42 nm in diameter) [2]. To classify this structural organization, a triangulation number can be used. The triangulation number (T) is a virology term describing the icosahedral packaging of viral capsid structural elements and is the quotient of dividing the number of subunits in a capsid by 60 [30]. Additionally, T can be explained as the number of subdivisions of each triangular facet of icosahedrons into identical equilateral triangles. The number of these triangles is equal to T. Following this classification, the Pyrococcus furiosus and Myxococcus xanthus encapsulins form icosahedrons with T equal to 3, Quasibacillus thermotolerans forms T = 4 icosahedrons, and Thermotoga maritima forms T = 1 icosahedron. The structure of an encapsulin shell protomer protein, like HK97 phage gp5 protomer, has three conserved domains: a peripheral domain (P), an axial domain (A), and an elongated loop (E) [2,6,14,30,31][2][6][14][30][31]. While the Pyrococcus furiosus and Myxococcus xanthus encapsulin protomer structures match the HK97 gp5 protomer structure well [13[13][24],24], only the A and E domains from the Thermotoga maritima encapsulin align well with those of the Pyrococcus furiosus and Myxococcus xanthus encapsulin protomer structures [2]. In particular, the E loop from Thermotoga maritima is shorter and rotated relative to the E loops of HK97 phage, Pyrococcus furiosus, and Myxococcus xanthus [2]. This allows the E loop to form a beta sheet with the E loop of a neighboring protomer, creating a tight interaction [2]. This may explain why the Thermotoga maritima encapsulin forms a T = 1 capsid, while the Pyrococcus furiosus and Myxococcus xanthus nanocompartments can form a larger T = 3 capsid.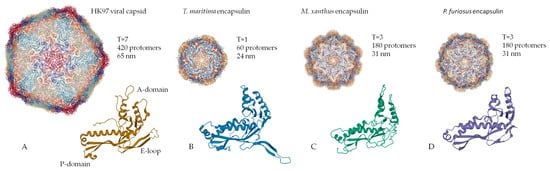
3. Cargo Proteins
The function of the nanocompartment is associated with the function of its protein cargo. The first insight into the mechanism of cargo encapsulation was obtained when the crystal structure of the Thermotoga maritima encapsulin was investigated in detail. X-ray crystallography revealed a small amount of extra electron density related to a hydrophobic pocket on the luminal surface of the encapsulin shell corresponding to a short (about 10 amino acid) C-terminal sequence of a ferritin-like protein (FLP), found next to the encapsulin shell gene in the Thermotoga maritima genome [2]. Bioinformatic analysis revealed that this C-terminal sequence is conserved in different species where the cargo protein genes and encapsulin shell genes are located together in a putative operon. Examples of such “predicted” cargo proteins include: FLP, DyP (dye-decolorizing peroxidase), hemerythrin, and rubrerythrin [2,20][2][20]. This C-terminal sequence, hereinafter termed the cargo loading peptide (CLP), is demonstrated to be sufficient for the loading of a cargo protein into the nanocompartment shell. The CLP coding sequence can be located at the 3′ or 5′ end of the cargo protein encoding gene (Figure 2A,B,D). Deletion of CLP from the cargo protein disrupts encapsulation, while fusion of the CLP with the C-terminus of heterologous proteins, such as green fluorescent protein or luciferase, is enough for loading [2,15,17,18,20,23][2][15][17][18][20][23]. Nevertheless, there are alternative models describing the interaction between shell proteins and cargo proteins. In some cases, such as the Pyrococcus furiosus encapsulins, CLP is absent but the gene encoding the shell is fused with the gene encoding the cargo protein, forming a single polypeptide (Figure 2C,E) [13]. In the encapsulins found in Firmicute bacteria, loading may occur partially via an N-terminus of CLP. Such a nanocompartment contains both an iron-mineralizing encapsulin-associated firmicute protein (IMEF) with C-terminal CLP and a ferredoxin protein with an N-terminal CLP [6].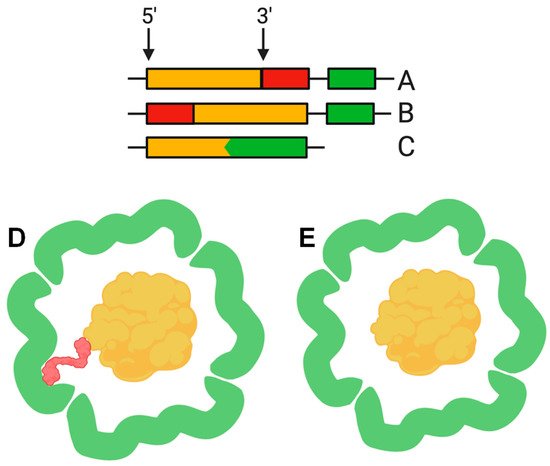
4. Nanocompartments and Nanotechnology
Over the last decade, a variety of nanoscale targeted delivery systems based on micelles [47,48][36][37], liposomes [49][38], inorganic [50][39] and polymer [51][40] nanoparticles, and protein compartments [52,53][41][42] have been developed. The small size and easy surface functionalization of these particles can increase the efficiency of drug delivery and accumulation compared with conventional nonencapsulated drugs. Such drugs are characterized by improved tissue penetration, longer circulation in the bloodstream, and reduced side effects [54][43]. Additionally, targeted drug delivery systems can play a significant role in the diagnosis of diseases by interacting with specific molecular markers expressed in a particular pathology. However, none of the existing nanoscale systems (liposomes, inorganic and polymer nanoparticles) can be encoded into the genome and self-assembled by the cell, whereas the use of encapsulins opens up prospects for this particular application. Encapsulins are mainly used as a diagnostic tool or as a drug delivery system. In various studies, Thermotoga maritima (Encap) encapsulins were used as a delivery system for both fluorescent probe and therapeutic drug when the nanocompartments were functionalized to bind to SP94, a hepatocellular carcinoma (HepG2) marker [14,55,56][14][44][45]. The Encap shell protein contains two cysteine residues (C123 and C197), while C123 is located on the outer surface of Encap, allowing for SP94 peptide conjugation to the encapsulin surface. Then, a fluorescent probe fluorescein-5-maleimide was added to the shell surface. Thus, the obtained nanodevice was able to specifically bind to cells and visualize SP94 simultaneously due to the presence of a fluorophore. It was further demonstrated that a prodrug aldoxorubicin ((6-maleimidocaproyl) doxorubicin hydrazone) could be loaded into the designed construct and released under acidic conditions inside tumor cells. The dose-dependent cytotoxicity of the drug against HepG2 cells was also demonstrated. Encapsulins can be successfully modified with photo-switchable fluorophores [57][46]. The Brevibacterium linens encapsulin was functionalized with spiropyran-based fluorophores (organic compounds with photochromic properties) via the carbodiimide method [58][47]. Upon irradiation with ultraviolet and visible light, spiropyrans are able to switch between their fluorescent merocyanine photo-isomer and nonradiative isomer, allowing them to “turn on” and “turn off” fluorescence [58][47]. The authors demonstrated that modified encapsulins carrying spiropyran molecules on their shell remained stable through at least five cycles of photo-switching. Modification of encapsulins with fluorescent dyes allows for investigating their intracellular fate and in vivo distribution, enabling a further understanding of their biological role and function. The possibility to switch the fluorescence on and off can significantly increase imaging resolution when using such super-resolution microscopy techniques as stochastic optical reconstruction microscopy (STORM) and photoactivated localization microscopy (PALM). In particular, spiropyran derivatives are widely used in live-cell imaging [59][48]. The sufficient brightness and versatile chemical modification of spiropyran-based compounds allow the attachment of multiple fluorophore molecules to the encapsulin shell [60][49]. The high sensitivity of spiropyrans to redox changes as well as to changes in temperature and pH make these molecular switchers an emerging tool in the development of fluorescent biosensors [61][50]. Moreover, due to their biocompatibility and ability to penetrate cells, encapsulin-based fluorescent probes may also facilitate cell labeling and tracking. Simultaneous encapsulin modification with fluorescent labels and targeting moieties might also lead to the creation of an encapsulin-based targeted imaging probe. In another study [36][51], Brevibacterium linens encapsulins loaded with teal fluorescent protein (TFP) were found to be successfully captured by J774 murine macrophages in cell culture in vitro, yielding the intense fluorescence of macrophages. In this case, encapsulins remain within the cytoplasm without entering the nucleus. The authors note that the selected fluorescent model cargo can be replaced by a therapeutic agent and an encapsulin-based platform can be used for its delivery. It was also shown that Thermotoga maritima encapsulins could be genetically modified to express the FcBP peptide (IgG-Fc domain-binding peptide) [62][52], which exhibits a high affinity to the rabbit IgG-Fc fragment [63][53]. Indeed, real-time quartz crystal microbalance (QCM) and surface plasmon resonance (SPR) analysis indicated that FcBP is displayed on the outer surface of encapsulin and is available for binding by rabbit IgG-Fc. While speaking about targeted delivery systems, it is worth mentioning that the encapsulin shell surface of Rhodococcus erythropolis N771, described above, can be successfully coated with polyethylene glycol (PEG). PEG is well known as a biocompatible modifier of drug carriers, making them undetectable by the cells of the monocytic macrophage system (mononuclear phagocyte system) and reducing aggregation. The authors also showed that PEGylation does not affect the self-assembly of the nanocompartment [21]. In one study, Thermotoga maritima encapsulins were used as carriers of the influenza A virus protein M2 ectodomain (M2e epitope) [64][54]. The M2 protein forms ion channels on the virion surface and is essential for the transport of viral ribonucleoprotein complexes into the cytoplasm of the host cell. The amino acid sequence of the M2 protein is highly conservative, and its immunogenicity during natural infection is quite low [65][55]. These properties make M2e a good broad-spectrum vaccine candidate. In this work, anti-M2e epitope specific antibodies were determined in mouse serum following immunization with the obtained constructs. Due to their size, protein compartments excellently mimic the defined intracellular environment and allow for studying the enzyme kinetics in more natural conditions [66,67][56][57]. It was shown that non-native proteins such as GFP and firefly luciferase (Luc) could be loaded into the encapsulin shell of Rhodococcus erythropolis [17]. Moreover, GFP retains its fluorescence properties, and luciferase showed enzymatic activity towards its substrate, luciferin. A similar study was conducted with Brevibacterium linens encapsulins. The C-terminal sequence of the native DyP cargo protein was fused to the C-terminus of TFP [68][58]. After confirming the structural integrity of the isolated nanocompartments, one encapsulin was found to contain 12 TFP molecules on average. Interestingly, the encapsulins can be disassembled into protein subunits at acidic pH, loaded with a cargo of interest due to the affinity to certain protein sequences of the membrane, and finally reassembled under neutral pH conditions. Thus, molecules/particles with a size exceeding the nanocompartment pore diameter can be loaded into the encapsulin cavity. Thus, the authors of [69][59] chose gold nanoparticles with a diameter of 13 ± 1 nm, coated with (11-mercaptoundecyl)-N,N,N-trimethylammonium bromide, as cargo. These nanoparticles were loaded into the Thermotoga maritima encapsulin cavity by partially replacing the nanoparticle-stabilizing ligand with CLP. This approach could be potentially applied for photothermal therapy of tumors, requiring the introduction of stable and biocompatible gold nanoparticles into the region of interest, followed by irradiation with the required wavelength and subsequent death of tumor cells, which are more sensitive to temperature increase compared to healthy cells.References
- Valdes-Stauber, N.; Scherer, S. Isolation and Characterization of Linocin M18, a Bacteriocin Produced by Brevibacterium Linens. Appl. Environ. Microbiol. 1994, 60, 3809–3814.
- Sutter, M.; Boehringer, D.; Gutmann, S.; Günther, S.; Prangishvili, D.; Loessner, M.J.; Stetter, K.O.; Weber-Ban, E.; Ban, N. Structural Basis of Enzyme Encapsulation into a Bacterial Nanocompartment. Nat. Struct. Mol. Biol. 2008, 15, 939–947.
- Giessen, T.W. Encapsulins: Microbial Nanocompartments with Applications in Biomedicine, Nanobiotechnology and Materials Science. Curr. Opin. Chem. Biol. 2016, 34, 1–10.
- Rosenkrands, I.; Rasmussen, P.B.; Carnio, M.; Jacobsen, S.; Theisen, M.; Andersen, P. Identification and Characterization of a 29-Kilodalton Protein from Mycobacterium Tuberculosis Culture Filtrate Recognized by Mouse Memory Effector Cells. Infect. Immun. 1998, 66, 2728–2735.
- Hicks, P.M.; Rinker, K.D.; Baker, J.R.; Kelly, R.M. Homomultimeric Protease in the Hyperthermophilic Bacterium Thermotoga Maritima Has Structural and Amino Acid Sequence Homology to Bacteriocins in Mesophilic Bacteria. FEBS Lett. 1998, 440, 393–398.
- Giessen, T.W.; Silver, P.A. Widespread Distribution of Encapsulin Nanocompartments Reveals Functional Diversity. Nat. Microbiol. 2017, 2, 17029.
- Winter, N.; Triccas, J.A.; Rivoire, B.; Pessolani, M.C.V.; Eiglmeier, K.; Lim, E.-M.; Hunter, S.W.; Brennan, P.J.; Britton, W.J. Characterization of the Gene Encoding the Immunodominant 35 KDa Protein of Mycobacterium Leprae. Mol. Microbiol. 1995, 16, 865–876.
- Triccas, J.A.; Roche, P.W.; Winter, N.; Feng, C.G.; Ruth Butlin, C.; Britton, W.J. A 35-Kilodalton Protein Is a Major Target of the Human Immune Response to Mycobacterium Leprae. Infect. Immun. 1996, 64, 5171–5177.
- Kawamoto, S.; Watanabe, M.; Saito, N.; Hesketh, A.; Vachalova, K.; Matsubara, K.; Ochi, K. Molecular and Functional Analyses of the Gene (EshA) Encoding the 52-Kilodalton Protein of Streptomyces Coelicolor A3(2) Required for Antibiotic Production. J. Bacteriol. 2001, 183, 6009–6016.
- Kwak, J.; McCue, L.A.; Trczianka, K.; Kendrick, K.E. Identification and Characterization of a Developmentally Regulated Protein, EshA, Required for Sporogenic Hyphal Branches in Streptomyces Griseus. J. Bacteriol. 2001, 183, 3004–3015.
- Saito, N.; Matsubara, K.; Watanabe, M.; Kato, F.; Ochi, K. Genetic and Biochemical Characterization of EshA, a Protein That Forms Large Multimers and Affects Developmental Processes in Streptomyces Griseus. J. Biol. Chem. 2003, 278, 5902–5911.
- Namba, K.; Hagiwara, K.; Tanaka, H.; Nakaishi, Y.; Chong, K.T.; Yamashita, E.; Armah, G.E.; Ono, Y.; Ishino, Y.; Omura, T.; et al. Expression and Molecular Characterization of Spherical Particles Derived from the Genome of the Hyperthermophilic Euryarchaeote Pyrococcus Furiosus. J. Biochem. 2005, 138, 193–199.
- Akita, F.; Chong, K.T.; Tanaka, H.; Yamashita, E.; Miyazaki, N.; Nakaishi, Y.; Suzuki, M.; Namba, K.; Ono, Y.; Tsukihara, T.; et al. The Crystal Structure of a Virus-like Particle from the Hyperthermophilic Archaeon Pyrococcus Furiosus Provides Insight into the Evolution of Viruses. J. Mol. Biol. 2007, 368, 1469–1483.
- Moon, H.; Lee, J.; Min, J.; Kang, S. Developing Genetically Engineered Encapsulin Protein Cage Nanoparticles as a Targeted Delivery Nanoplatform. Biomacromolecules 2014, 15, 3794–3801.
- Rurup, W.F.; Snijder, J.; Koay, M.S.T.; Heck, A.J.R.; Cornelissen, J.J.L.M. Self-Sorting of Foreign Proteins in a Bacterial Nanocompartment. J. Am. Chem. Soc. 2014, 136, 3828–3832.
- Snijder, J.; Van De Waterbeemd, M.; Damoc, E.; Denisov, E.; Grinfeld, D.; Bennett, A.; Agbandje-Mckenna, M.; Makarov, A.; Heck, A.J.R. Defining the Stoichiometry and Cargo Load of Viral and Bacterial Nanoparticles by Orbitrap Mass Spectrometry. J. Am. Chem. Soc. 2014, 136, 7295–7299.
- Tamura, A.; Fukutani, Y.; Takami, T.; Fujii, M.; Nakaguchi, Y.; Murakami, Y.; Noguchi, K.; Yohda, M.; Odaka, M. Packaging Guest Proteins into the Encapsulin Nanocompartment from Rhodococcus Erythropolis N771. Biotechnol. Bioeng. 2015, 112, 13–20.
- Cassidy-Amstutz, C.; Oltrogge, L.; Going, C.C.; Lee, A.; Teng, P.; Quintanilla, D.; East-Seletsky, A.; Williams, E.R.; Savage, D.F. Identification of a Minimal Peptide Tag for in Vivo and in Vitro Loading of Encapsulin. Biochemistry 2016, 55, 3461–3468.
- Choi, B.; Moon, H.; Hong, S.J.; Shin, C.; Do, Y.; Ryu, S.; Kang, S. Effective Delivery of Antigen-Encapsulin Nanoparticle Fusions to Dendritic Cells Leads to Antigen-Specific Cytotoxic T Cell Activation and Tumor Rejection. ACS Nano 2016, 10, 7339–7350.
- Giessen, T.W.; Silver, P.A. Converting a Natural Protein Compartment into a Nanofactory for the Size-Constrained Synthesis of Antimicrobial Silver Nanoparticles. ACS Synth. Biol. 2016, 5, 1497–1504.
- Sonotaki, S.; Takami, T.; Noguchi, K.; Odaka, M.; Yohda, M.; Murakami, Y. Successful PEGylation of Hollow Encapsulin Nanoparticles from: Rhodococcus Erythropolis N771 without Affecting Their Disassembly and Reassembly Properties. Biomater. Sci. 2017, 5, 1082–1089.
- Rahmanpour, R.; Bugg, T.D.H. Assembly in Vitro of Rhodococcus Jostii RHA1 Encapsulin and Peroxidase DypB to Form a Nanocompartment. FEBS J. 2013, 280, 2097–2104.
- Contreras, H.; Joens, M.S.; McMath, L.M.; Le, V.P.; Tullius, M.V.; Kimmey, J.M.; Bionghi, N.; Horwitz, M.A.; Fitzpatrick, J.A.J.; Goulding, C.W. Characterization of a Mycobacterium Tuberculosis Nanocompartment and Its Potential Cargo Proteins. J. Biol. Chem. 2014, 289, 18279–18289.
- McHugh, C.A.; Fontana, J.; Nemecek, D.; Cheng, N.; Aksyuk, A.A.; Heymann, J.B.; Winkler, D.C.; Lam, A.S.; Wall, J.S.; Steven, A.C.; et al. A Virus Capsid-like Nanocompartment That Stores Iron and Protects Bacteria from Oxidative Stress. EMBO J. 2014, 33, 1896–1911.
- He, D.; Hughes, S.; Vanden-Hehir, S.; Georgiev, A.; Altenbach, K.; Tarrant, E.; Mackay, C.L.; Waldron, K.J.; Clarke, D.J.; Marles-Wright, J. Structural Characterization of Encapsulated Ferritin Provides Insight into Iron Storage in Bacterial Nanocompartments. Elife 2016, 5, e18972.
- Wikoff, W.R.; Liljas, L.; Duda, R.L.; Tsuruta, H.; Hendrix, R.W.; Johnson, J.E. Topologically Linked Protein Rings in the Bacteriophage HK97 Capsid. Science (80-) 2000, 289, 2129–2133.
- Fokine, A.; Leiman, P.G.; Shneider, M.M.; Ahvazi, B.; Boeshans, K.M.; Steven, A.C.; Black, L.W.; Mesyanzhinov, V.V.; Rossmann, M.G. Structural and Functional Similarities between the Capsid Proteins of Bacteriophages T4 and HK97 Point to a Common Ancestry. Proc. Natl. Acad. Sci. USA 2005, 102, 7163–7168.
- Baker, M.L.; Jiang, W.; Rixon, F.J.; Chiu, W. Common Ancestry of Herpesviruses and Tailed DNA Bacteriophages. J. Virol. 2005, 79, 14967–14970.
- Pietilä, M.K.; Laurinmäki, P.; Russell, D.A.; Ko, C.C.; Jacobs-Sera, D.; Hendrix, R.W.; Bamford, D.H.; Butcher, S.J. Structure of the Archaeal Head-Tailed Virus HSTV-1 Completes the HK97 Fold Story. Proc. Natl. Acad. Sci. USA 2013, 110, 10604–10609.
- CASPAR, D.L.; KLUG, A. Physical Principles in the Construction of Regular Viruses. Cold Spring Harb. Symp. Quant. Biol. 1962, 27, 1–24.
- Giessen, T.W.; Orlando, B.J.; Verdegaal, A.A.; Chambers, M.G.; Gardener, J.; Bell, D.C.; Birrane, G.; Liao, M.; Silver, P.A. Large Protein Organelles Form a New Iron Sequestration System with High Storage Capacity. Elife 2019, 8, e46070.
- Bamford, D.H.; Grimes, J.M.; Stuart, D.I. What Does Structure Tell Us about Virus Evolution? Curr. Opin. Struct. Biol. 2005, 15, 655–663.
- Heinemann, J.; Maaty, W.S.; Gauss, G.H.; Akkaladevi, N.; Brumfield, S.K.; Rayaprolu, V.; Young, M.J.; Lawrence, C.M.; Bothner, B. Fossil Record of an Archaeal HK97-like Provirus. Virology 2011, 417, 362–368.
- Williams, E.M.; Jung, S.M.; Coffman, J.L.; Lutz, S. Pore Engineering for Enhanced Mass Transport in Encapsulin Nanocompartments. ACS Synth. Biol. 2018, 7, 2514–2517.
- Snijder, J.; Kononova, O.; Barbu, I.M.; Uetrecht, C.; Rurup, W.F.; Burnley, R.J.; Koay, M.S.T.; Cornelissen, J.J.L.M.; Roos, W.H.; Barsegov, V.; et al. Assembly and Mechanical Properties of the Cargo-Free and Cargo-Loaded Bacterial Nanocompartment Encapsulin. Biomacromolecules 2016, 17, 2522–2529.
- Gong, J.; Chen, M.; Zheng, Y.; Wang, S.; Wang, Y. Polymeric Micelles Drug Delivery System in Oncology. J. Control. Release 2012, 159, 312–323.
- Rösler, A.; Vandermeulen, G.W.M.; Klok, H.A. Advanced Drug Delivery Devices via Self-Assembly of Amphiphilic Block Copolymers. Adv. Drug Deliv. Rev. 2012, 64, 270–279.
- Allen, T.M.; Cullis, P.R. Liposomal Drug Delivery Systems: From Concept to Clinical Applications. Adv. Drug Deliv. Rev. 2013, 65, 36–48.
- Wang, A.Z.; Langer, R.; Farokhzad, O.C. Nanoparticle Delivery of Cancer Drugs. Annu. Rev. Med. 2012, 63, 185–198.
- Haag, R.; Kratz, F. Polymer Therapeutics: Concepts and Applications. Angew. Chem. Int. Ed. 2006, 13, 1198–1215.
- Ma, Y.; Nolte, R.J.M.; Cornelissen, J.J.L.M. Virus-Based Nanocarriers for Drug Delivery. Adv. Drug Deliv. Rev. 2012, 64, 811–825.
- MaHam, A.; Tang, Z.; Wu, H.; Wang, J.; Lin, Y. Protein-Based Nanomedicine Platforms for Drug Delivery. Small 2009, 5, 1706–1721.
- Brigger, I.; Dubernet, C.; Couvreur, P. Nanoparticles in Cancer Therapy and Diagnosis. Adv. Drug Deliv. Rev. 2002, 54, 631–651.
- Toita, R.; Murata, M.; Tabata, S.; Abe, K.; Narahara, S.; Piao, J.S.; Kang, J.H.; Hashizume, M. Development of Human Hepatocellular Carcinoma Cell-Targeted Protein Cages. Bioconjug. Chem. 2012, 23, 1494–1501.
- Toita, R.; Murata, M.; Abe, K.; Narahara, S.; Piao, J.S.; Kang, J.H.; Hashizume, M. A Nanocarrier Based on a Genetically Engineered Protein Cage to Deliver Doxorubicin to Human Hepatocellular Carcinoma Cells. Chem. Commun. 2013, 49, 7442–7444.
- Putri, R.M.; Fredy, J.W.; Cornelissen, J.J.L.M.; Koay, M.S.T.; Katsonis, N. Labelling Bacterial Nanocages with Photo-Switchable Fluorophores. Chem. Phys. Chem. 2016, 1815–1818.
- Klajn, R. Spiropyran-Based Dynamic Materials. Chem. Soc. Rev. 2014, 43, 148–184.
- Fernández-Suárez, M.; Ting, A.Y. Fluorescent Probes for Super-Resolution Imaging in Living Cells. Nat. Rev. Mol. Cell Boil. 2008, 9, 929–943.
- Fihey, A.; Perrier, A.; Browne, W.R.; Jacquemin, D. Multiphotochromic Molecular Systems. Chem. Soc. Rev. 2015, 44, 3719–3759.
- Kortekaas, L.; Browne, W.R. The Evolution of Spiropyran: Fundamentals and Progress of an Extraordinarily Versatile Photochrome. Chem. Soc. Rev. 2019, 48, 3406–3424.
- Putri, R.M.; Allende-Ballestero, C.; Luque, D.; Klem, R.; Rousou, K.A.; Liu, A.; Traulsen, C.H.H.; Rurup, W.F.; Koay, M.S.T.; Castón, J.R.; et al. Structural Characterization of Native and Modified Encapsulins as Nanoplatforms for in Vitro Catalysis and Cellular Uptake. ACS Nano 2017, 11, 12796–12804.
- Moon, H.; Lee, J.; Kim, H.; Heo, S.; Min, J.; Kang, S. Genetically Engineering Encapsulin Protein Cage Nanoparticle as a SCC-7 Cell Targeting Optical Nanoprobe. Biomater. Res. 2014, 18, 21.
- Jung, Y.; Kang, H.J.; Lee, J.M.; Jung, S.O.; Yun, W.S.; Chung, S.J.; Chung, B.H. Controlled Antibody Immobilization onto Immunoanalytical Platforms by Synthetic Peptide. Anal. Biochem. 2008, 374, 99–105.
- Lagoutte, P.; Mignon, C.; Stadthagen, G.; Potisopon, S.; Donnat, S.; Mast, J.; Lugari, A.; Werle, B. Simultaneous Surface Display and Cargo Loading of Encapsulin Nanocompartments and Their Use for Rational Vaccine Design. Vaccine 2018, 36, 3622–3628.
- Cho, K.J.; Schepens, B.; Seok, J.H.; Kim, S.; Roose, K.; Lee, J.-H.; Gallardo, R.; Van Hamme, E.; Schymkowitz, J.; Rousseau, F.; et al. Structure of the Extracellular Domain of Matrix Protein 2 of Influenza A Virus in Complex with a Protective Monoclonal Antibody. J. Virol. 2015, 89, 3700–3711.
- Maity, B.; Fujita, K.; Ueno, T. Use of the Confined Spaces of Apo-Ferritin and Virus Capsids as Nanoreactors for Catalytic Reactions. Curr. Opin. Chem. Boil. 2015, 25, 88–97.
- Safont-Sempere, M.M.; Fernández, G.; Würthner, F. Self-Sorting Phenomena in Complex Supramolecular Systems. Chem. Rev. 2011, 111, 5784–5814.
- Ai, H.W.; Henderson, J.N.; Remington, S.J.; Campbell, R.E. Directed Evolution of a Monomeric, Bright and Photostable Version of Clavularia Cyan Fluorescent Protein: Structural Characterization and Applications in Fluorescence Imaging. Biochem. J. 2006, 400, 531–540.
- Künzle, M.; Mangler, J.; Lach, M.; Beck, T. Peptide-Directed Encapsulation of Inorganic Nanoparticles into Protein Containers. Nanoscale 2018, 10, 22917–22926.
