Lymphocyte migration to and sequestration in specific microenvironments plays a crucial role in their differentiation and survival. Lymphocyte trafficking and homing is tightly regulated by signaling pathways and is mediated by cytokines, chemokines, cytokine/chemokine receptors, and adhesion molecules. The production of cytokines and chemokines is largely controlled by transcription factors in the context of a specific epigenetic landscape. These regulatory factors are strongly inter-connected, and they influence the gene expression pattern in lymphocytes, promoting processes such as cell survival. The epigenetic status of the genome plays a key role in regulating gene expression during many key biological processes and it is becoming more evident that dysregulation of epigenetic mechanisms contributes to cancer initiation, progression and drug resistance. Here, we review the signaling pathways that regulate lymphoma cell migration and adhesion with a focus on Mantle cell lymphoma and highlight the fundamental role of epigenetic mechanisms in integrating signals at the level of gene expression throughout the genome.
- mantle cell lymphoma
- tumor microenvironment
- lymphoma trafficking
- epigenetic status
- gene expression
- epigenetic landscape
- signaling pathway
- cytokine and chemokine
- adhesion molecules
- Histone modifications
1. Epigenetic rRegulation of tumor-mTumor-Microenvironment 1. Introduction
The tumor microenvironment, which includes various types of cells, is a location where tumor cells continuously interact with surrounding normal cells and extracellular molecules. In B-cell lymphomas, interaction of tumor cells with stromal cells in the tumor microenvironment promotes tumor cell growth, proliferation, survival and drug resistance [1,2,3]. The interactions between malignant B-cells and tumor microenvironment components alter the gene expression profiles of tumor cells in order to favor the survival of malignant B-cells [4,5,6].
Mantle cell lymphoma (MCL) is a rare and largely incurable B-cell non-Hodgkin lymphoma (NHL) accounting for 5–10% of all lymphomas [7]. This results in cyclin D1 overexpression and leads to aberrant cell cycle progression [8,9]. These studies demonstrated several recurrent mutations, includingATM,TP53andMLL2,but they also uncovered a number of novel and infrequent mutations in patients, which suggests divergent clonal evolution pattern in MCL cells [24]. As tumor cells divide, mutations occur randomly, resulting in multiple sub-clonal populations with different phenotypes within the same tumor, known as intra-tumor heterogeneity, which makes tumor more prone to recurrence and drug resistance.
Distinct from mutations, altered epigenetic status, i.e., chromosomal conditions that influence gene expression without changing the primary DNA sequence, plays an important role in the development of B-cell lymphomas [25,26,27]. Variable epigenetic components throughout the genome that influence gene expression include DNA methylation and patterns of histone modification, which alter chromatin structure, DNA accessibility and gene expression patterns [28]. The combination of genetic mutations and epigenetic changes induced by internal and external factors leads to constitutive activation of proto-oncogenes and the loss of tumor-suppressor gene activity that ultimately causes tumorigenesis [25,29,30]. Comprehensive methylation profiling by genome-wide array, comparing DNA methylation changes in MCL patients with normal B-cells, has demonstrated highly heterogenous methylation profiles across MCL patient samples [31].
In addition to the role of intrinsic abnormalities in tumor cells, an essential role of extrinsic factors, such as the tumor microenvironment, in tumor growth and development of drug resistance in B-cell lymphomas is widely accepted [35,36,37]. Interactions between tumor cells and microenvironment components trigger activation of a cascade of signals that travel to the nucleus, integrate with the specific epigenetic landscape of the tumor cell and thereafter cause specific alterations in gene expression. Crosstalk between tumor cells and microenvironment components promotes anti-tumor immune responses in B-cell lymphomas [46]. Although the role of the tumor microenvironment in the regulation of cellular transduction pathways in tumor cells has already been described [6,54,55], much less is known about the interaction between cell signaling events and the specific epigenetic landscape of the cells that causes cell-type-specific effects on chromatin structure and thereby gene expression.
Here, we review current evidence indicating that epigenetic alterations contribute to the development of NHLs including MCL. Recent investigations have identified mutations in epigenetic regulators that cause epigenetic changes and thereby lead to abnormal gene activity in lymphocytes [30,56,57]. However, epigenetic alterations in lymphocytes have also been observed in the absence of mutations in genes encoding proteins important for epigenetic regulation, suggesting that the tumor microenvironment can induce epigenetic dysregulation in lymphoma cells. We further propose a possible model where somatic mutations and the tumor microenvironment interact in order to shape the epigenetic landscape of lymphoma cells, which induces transcriptional changes in tumor cells, enabling their proliferation and survival.
2. Epigenetic Alteration in MCL
Chromatin structure and organization play a crucial role in regulating transcription by modulating DNA accessibility to the transcriptional machinery and transcription factors. The establishment and maintenance of distinct chromatin domains play an important role by establishing the pre-conditions that determine the extent to which individual genes can be regulated by transcription factors [59]. Multiple mechanisms affecting chromatin organization, including DNA methylation, ATP-dependent chromatin remodeling and post-translational modification of histones, are required to ensure proper cellular functions. Different modifications of histone proteins are associated with discrete chromatin states that may be activated or repressed and impact a wide variety of cellular processes including transcription, DNA replication and DNA repair [60].
Currently known disease-causing epigenetic changes in B-cell lymphomas are predominantly induced by somatic mutations in epigenetic regulators, which have been identified by genome-wide screens and include key players involved in DNA methylation, histone modification and chromatin organization [32,57,61,62]. Here, we will briefly discuss mutations that modulate the epigenetic landscape of MCL as well as some other lymphomas, resulting in an abnormal transcriptional programming (Figure 1). A list of mutations known to be involved in epigenetic alterations in MCL is presented in Table 1.
| Dysregulated Epigenetic Mark in MCL | Gene/Genes Mediating Epigenetic Dysregulation in MCL | Type of Dysregulation | Target Gene/Genes Affected by Epigenetic Dysregulation | Putative Role in MCL | Reference |
|---|---|---|---|---|---|
| DNA methylation | - | DNA hypomethylation at promoter region of target gens | Mediating the expression of SOX11 | Promotes oncogenic cell proliferation | [63] |
| DNA methylation | DNMT1 upregulation | Global DNA hypermethylation | Abnormal expression of β-catenin which upregulates the expression of c-MYC and MMP7 (Matrix Metallopeptidase 7) Reduced expression of tumor suppressor gene; PARG1 |
Promotes tumor cell proliferation and survival | [64,65] |
| Histone H3K27acetylation | Mutation in the component of SWI-SNF complex (SMARCA4) | Abnormal histone H3K27 acetylation and chromatin accessibility at the promoter/enhancer region of target genes | Reduced chromatin accessibility at promoter region of transcription factor ATF3 (negative regulator of anti-apoptotic gene BCL-xL) | MCL cell survival and drug resistance | [32,66,67] |
| Global histone acetylation | Abnormal activity of HDACs, Class I, II, e.g., HDAC8 | Enhanced HDAC (Histone deacetylase) activity leading to abnormal histone acetylation and chromatin accessibility | Reduced transcription of pro-apoptotic genes (BIM, BMF) Enhanced expression of c-MYC and PLK1 |
Inhibits apoptosis and promotes tumor cell proliferation and survival | [68,69,70,71] |
| Histone H3K36me3 | Gain of function mutation in histone methyltransferase WHSC1 (MMSET) | Enhanced H3K36me3 levels | Enhanced expression of cell cycle regulators | Promotes tumor cell proliferation | [72,73] |
| Histone H3K4 methylation | Loss of function mutation in histone methyltransferase MLL2 (KMT2D) | Diminished H3K4 methylation levels | Functional consequences in MCL are not well understood Contributes to genome instability and transcriptional stress |
Disturbs the expression of genes that sustain proliferation and cell survival | [32,74,75,76] |
| H3K27me3 | EZH2 upregulation | Enhanced H3K27me3 levels | Repressed expression of CDKN2B, HOX genes | Promotes MCL cell growth | [77,78] |
| H3K27me3 | KDM6B histone demethylase | Enhanced KDM6B levels, reduces H3K27me3 at promoter region of target genes | Functional consequences in MCL are not well understood In other B-cell malignancies target NF-κB subunits and target genes |
Promoters tumor cells survival and drug resistance | [79,80,81] |
Extensive perturbation of cytosine methylation patterns is a hallmark of B-cell malignancies and occurs in specific patterns that can be distinguished in different subtypes of NHLs [63]. Analysis of the genome-wide DNA methylation profile in MCL patients revealed significantly different promoter methylation patterns between MCL patient samples and normal B-cells [63]. who showed that DNA methyltransferase-1 (DNMT-1) is upregulated in MCL, causing global disruption of DNA methylation patterns [64]. In general, abnormal DNA methylation of tumor suppressor genes suppresses their transcription levels, and this is a common phenomenon observed in cancer.
Nucleosomes play an important role in regulating higher-order chromatin structure. Chromatin remodeling enzymes use ATP to alter the structure and/or position of nucleosomes and thereby regulate the accessibility of DNA to transcription factors [84]. In eukaryotic cells, a wide variety of chromatin-remodeling complexes are present to remodel chromatin via different mechanisms [85]. For example, the SWI/SNF family of chromatin remodeling enzymes globally moves or evicts nucleosomes in order to create open chromatin regions, leading to transcriptional activation of genes throughout the genome [86].
Mutations in genes encoding proteins involved in chromatin remodeling have been identified in different types of cancer. SMARCA4, a central component of the SWI/SNF chromatin-remodeling complex, regulates transcription via remodeling nucleosome positions. MCL patients with aSMARCA4mutation are resistant to combined therapy with Ibrutinib (BTK inhibitor) and Venetoclax (BCL-2 inhibitor) The mutation inSMARCA4affects chromatin accessibility and reduces expression of the transcription factor, ATF3, which negatively regulates transcription of the anti-apoptotic geneBCL-xL,
In general, histone acetylation is associated with active transcription. CBP, known as (P300/CBP), acetylates histone and non-histone proteins in order to activate transcription [87]. In contrast, histone deacetylases (HDACs) remove acetyl groups from hyperacetylated histones in order to suppress gene transcription [88]. In addition to histone deacetylation, HDACs also regulate the acetylation status of a variety of non-histone proteins [89].
HDAC inhibitors (HDACi) are a class of chemical compounds that increase histone acetylation and restore the balance between pro- and anti-apoptotic proteins and induce apoptosis in tumor cells. Inhibition of HDACs induced cell death in MCL cell lines by enhancing histone acetylation at the promoter region of pro-apoptotic genes including (BIMandBMF), which induced their transcription [68]. In addition, Vorinostat in combination with Palbociclib (selective inhibitor of cyclin-dependent kinase 4/6 involved in cell cycle progression) reduced MCL cell growth and induced apoptosis significantly by increasing histone H3 acetylation and inhibiting BCL-2 family members. Overexpression of BCL-2 family proteins attenuates apoptosis in tumor cells [69].
Another HDACi (PCI-24781) enhanced acetylation of histone H3 at the promoter region of the gene encoding P21 (CDKN1A), causing increased chromatin accessibility for binding of transcription factors [70]. Moreover, PCI-24781 significantly downregulated the expression of NF-κB subunits (NF-κB1 and REL B) and NF-κB This suggests a role for histone deacetylation in the regulation of NF-κB activity, which remains poorly understood. A combination of PCI-24781 and bortezomib (a proteasome inhibitor) synergistically suppressed NF-κB and induced apoptosis in lymphoma cells [70].
HDAC3 has been implicated as a regulator of PD-L1 expression in B-cell lymphomas [94]. Inhibition of HDAC3 reversed microenvironment-mediated immunosuppression and resulted in better clinical response to PD-L1 blockage [94]. In Chronic lymphocytic leukemia (CLL), the combination of HDAC6 inhibitor (ACY-738) with PD-L1 blockage using monoclonal antibodies significantly increased anti-tumor efficiency of T-cells and reduced tumor burden [95]. Thus, the combination of HDAC inhibitors and checkpoint blockage can help to overcome immunotherapy resistance.
Post-translational modification of histone proteins by methylation is an important and common modification that influences many cellular processes. The major methylation sites on histone proteins are lysine and arginine residues and methylation of non-histone proteins is also frequently observed [96,97]. Histone methylation is a dynamic process, and methyl groups can be removed from histone proteins by histone demethylases [98]. The abnormal expression of various methyltransferases and demethylases has been reported in many tumor types, suggesting that the level and degree of histone methylation contributes to tumorigenesis and that the enzymes involved provide promising targets for anti-cancer treatment [79,99,100].
WHSC1 (also known as MMSET or NSD2) is a specific histone methyltransferase, responsible for histone H3 methylation at lysine 36, and it is mutated in a subgroup of B-cell malignancies including MCL [72,73]. Alterations in histone methylation status can switch a gene from an active to a repressed state or vice versa. As a result of altered WHSC1 levels, the expression of genes positively involved in proliferation and cell-cycle progression increases, suggesting thatWHSC1functions as an oncogene favoring tumor cell growth and proliferation.
While H3K4me3 is associated with the activation of gene expression, H3K27me3 is a mark for repressed genes [104,105]. The promoter region is generally uniquely marked with either H3K4me3 or H3K27me3, except in the case of bivalent chromatin at the promoter region of developmental regulatory genes [104,105,106]. Bivalent chromatin is marked with histone modifications associated with both gene activation and repression and keeps repressed genes poised and ready for rapid activation [106]. Bernhart S et al., using ChIP-seq (chromatin immunoprecipitation combined with high-throughput sequencing), demonstrated that chromatin structure at bivalent promoters in cancer cells (both in solid and hematological tumors) is disrupted, resulting in deregulation of gene silencing and activation of poised genes [107].
MLL2has been identified as one of the commonly mutated genes in NHLs including Diffuse large B-cell lymphoma (DLBCL) and MCL [66,67,74]. In DLBCL, mutation ofMLL2produces a truncatedMLL2protein that lacks the SET domain (required for its methyltransferase activity) [74]. Although the role ofMLL2in MCL progression is not well understood,MLL2loss-of-function mutations are known to diminish H3K4 methylation and drive tumor cell growth and proliferation, suggesting thatMLL2functions as a tumor suppressor gene [75,108]. Interestingly, a recent study has provided evidence for the role ofMLL2in maintaining genomic stability, since the loss ofMLL2caused chromosomal aberrations and accumulation of mutations [76].
The multi-protein polycomb complexes (PRCs) promote transcriptional repression by disturbing nucleosome occupancy and mediating H3K27 methylation. All three forms of H3K27 methylation are mutually exclusive and are present at distinct genomic regions. Trimethylated H3K27 (H3K27me3) is present at promoter regions of silenced genes and is associated with transcriptional repression [112]. To preserve chromatin structure and prolong gene silencing, H3K27me3 recruits PRC complexes to nucleosomes of the nascent DNA strand during DNA replication in order to maintain H3K27me3 levels [113].
One of the most frequent genetic alterations observed in germinal center B-cell (GCB) lymphomas such as Follicular lymphoma (FL) and DLBCL is mutations ofEZH2that affect the SET domain. EZH2is essential for normal B-cell development and rearrangement of the immunoglobulin heavy chain (IGHV) [114]. Moreover,EZH2is required for the differentiation of germinal B-cells into memory cells through the establishment and maintenance of bivalent modifications at the promoter region of key regulatory genes [115].
In addition to mutation, other genetic lesions affectingEZH2, including chromosomal gain or loss and DNA hypermethylation, have been identified in B-cell malignancies The work of Demosthenous et al. showed that deregulation of PRC2/EZH2 in MCL cell lines mediates epigenetic silencing of theCDKN2BCDK inhibitor gene to promote MCL cell proliferation and survival [77]. In addition, it has been shown that EZH2 overexpression in MCL cell lines and primary cells facilitates recruitment of the DNA methylation machinery, enabling more stable and long-term repression of HOX genes [78]. Further studies are required to develop strategies to specifically alter H3K27me3 levels in tumor cells.
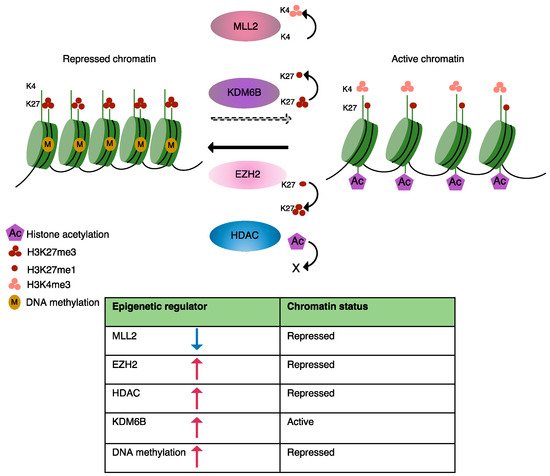
Post-translational modification of histone H3 at lysine 27 has an essential role in both transcriptional repression and activation. Studies have shown that both increase and decrease in the activity of enzymes regulating H3K27me3 levels are associated with carcinogenesis [61,80]. Despite the opposing activities of EZH2 and the KDM6B demethylase in affecting H3K27me3 levels, both these enzymes have been implicated in both lymphoma development and drug resistance.
Histone demethylase JMJD3 removes di- and tri-methyl marks from H3K27 and reverses PRC2/EZH2-mediated transcription repression [119,120]. In contrast, KDM6B overexpression has been observed in Hodgkin’s lymphomas (HLs), DLBCL, Multiple myeloma (MM) and Acute myeloid leukemia (AML) KDM6B is required for maintaining hematopoietic stem cell (HSC) self-renewal ability and repopulation capacity and plays an important role in the differentiation of memory B-cells [122,124].
In MM, KDM6B regulates the expression of key components of the MAP Kinase pathway in a demethylase-activity-independent manner in order to enhance tumor cell survival, whereas in DLBCL, KDM6B controls BCL6 expression and protein levels in a fashion that requires the demethylase activity in order to facilitate tumor cell proliferation [79,80]. KDM6B has been identified as a new therapeutic target in B-cell malignancies, and its selective inhibitor GSK-J4 has been shown to counteract cell proliferation in different tumor types such as AML, MM and DLBCL [80,123].
KDM6B overexpression can antagonize EZH2-mediated repression [125] of tumor suppressor genes in solid tumors by reducing H3K27me3 levels, while inhibition of KDM6B boosts apoptotic response to PI3K/AKT inhibitor treatment. This suggests that KDM6B can act as a tumor suppressor gene or an oncogene depending on cellular context [126].
3. Tumor-Microenvironment Mediated Epigenetic Alterations
As discussed above, cancer-associated mutations that affect the epigenetic machinery can lead to imbalances that change the epigenetic landscape in cancer cells and its effect on gene expression programs. Regulation of gene expression in response to signaling pathways requires post-translational modifications of histones and nucleosome rearrangement, which promotes chromatin reorganization, leading to activation or repression of transcription at particular loci throughout the genome. Interactions between malignant B-cells and stromal cells activate multiple signaling pathways, including PI3K/AKT and NF-κB, that function as signal transducers for cell-surface receptors, particularly receptor tyrosine kinases, in order to alter cellular function [6]. Here, we focus on some examples of key tumor microenvironment-mediated signaling pathways that have a direct impact on the chromatin state of tumor cells.
The PI3K/AKT pathway plays a key role in many cellular mechanisms and is often dysregulated in malignant B-cells [127]. The PI3K/AKT pathway regulates essential cellular functions including growth, proliferation, metabolism, differentiation and motility in normal and malignant B-cells [128]. to generate phosphatidylinositol-3,4,5-trisphosphate (PIP3), which in turn acts as a second messenger to assemble and activate downstream signaling complexes, including protein kinase B, also known as AKT [129]. Phosphorylated AKT mediates phosphorylation of different proteins, including the mammalian target of rapamycin (mTOR), and the IκB inhibitor of NF
Adhesion of MCL cells to stromal cells in the tumor microenvironment activates the PI3K pathway, which transmits signals from cell membrane receptors, such as the B-cell receptor and CXCR4 (chemokine receptor), to the nucleus, leading to transcriptional regulation of genes involved in migration, proliferation and survival [45,127,132,133]. AKT-mediated phosphorylation of P300/CBP facilitates recruitment of the transcription machinery at the promoter region of genes involved in the survival of MCL and CLL cells [127,136]. Expression levels of CXCR4 are regulated by the transcription factor, FOXO1, and acetylation of histones by P300/CBP is a prerequisite for FOXO1/P300-mediated recruitment of transcription machinery at the promoter regions of important target genes [138,139]. Zhou XR et al. reported that a P300/CBP inhibitor (A-485) overcomes resistance to Idelalisib in MCL by reducing histone acetylation at the promoter region of receptor tyrosine kinase (RTK) genes, thereby, reducing their transcriptional upregulation by an independent mechanism [140].
In mammals, DNA methylation is performed by DNMT1 and DNMT3, which are functionally and structurally distinct [142]. In general, maintenance of DNA methylation activity is essential for the preservation of genomic integrity and inhibition of inappropriate gene activation. Abnormal activation of PI3K/AKT might contribute to tumor cell growth by favoring DNA hypermethylation and chromatin compaction at the promoter region of specific target genes, thereby suppressing their expression [141]. Accumulating evidence demonstrates a role for DNMT1 in malignant progression of NHLs including DLBCL and MCL by enhancing cell cycle progression [64,143,144].
In addition, PI3K/AKT is involved in regulating histone methylation levels. AKT interacts with and phosphorylates EZH2 at Serine 21 and suppresses its methyltransferase activity, which results in reduced H3K27me3 levels at the promoter regions of target genes, thus disrupting gene silencing [145]. A Serine to Alanine substitution at the phosphorylation site increases EZH2 association with H3 [145].
Tumor microenvironment-mediated upregulation of EZH2 has been reported in various types of NHL [146,147]. In a subset of CLL cells, signals from the microenvironment result in pronounced upregulation of EZH2 mRNA and protein levels, which induces anti-apoptotic responses and enhances cell viability [146]. Moreover, dysregulation of EZH2 levels in CLL results in intra-tumor heterogeneity contributing to drug resistance [148].
NF-κB binds to the promoter and/or enhancer region of its target genes, which are involved in stress responses, cell proliferation and apoptosis, and regulates their transcription [150]. The NF–κB The NF-κB signaling pathway is divided into separate canonical and non-canonical pathways that regulate distinct gene expression programs. Disruption of the NF-κB
Activation of NF-κB in response to cytokines and chemokines in tumor microenvironments promotes upregulation of EZH2 in leukemia and lymphoma cell lines [154]. Natoli G et al. showed that exposure of macrophages to different inflammatory cytokines elevated KDM6B In a recent study from our lab, we showed that stromal cell adhesion mediates overexpression of KDM6B in MCL cell lines, which could potentially lead to the removal of the H3K27me3 mark from the promoter regions of genes encoding NF-κB subunits and thus activate their expression and nuclear localization. activity mediates transcription of migration and adhesion-related genes and promotes tumor cell survival.
4. Targeting PI3K in MCL
Given the crucial role of PI3K/AKT in lymphoma development and survival, different compounds have been identified that inhibit the PI3K activity. Idelalisib (formerly known as GS-1101 and CAL-101), the first class of PI3K inhibitors, has been used as a single-agent monotherapy or in combination with other inhibitors/agents to treat refractory or relapsed MCL and CLL patients [155]. Idelalisib inhibits MCL and CLL cell migration toward CXCL12 gradients, promotes egression of malignant B-cells from lymph nodes and induces apoptosis of malignant B-cells [155,156,157].
Combined PI3K and HDAC inhibitors have demonstrated significant anti-tumor effects in MCL and DLBCL [71,158]. CUDC-907 (dual PI3K and HDAC inhibitor) inhibits growth and proliferation of both Ibrutinib-sensitive and Ibrutinib-resistant MCL primary cells in vitro and induces apoptosis and cell cycle arrest through downregulation of PLK1, polo-like kinase 1, a key regulator of mitosis [71]. Moreover, CUDC-907 downregulated MYC in DLBCL and showed a promising anti-tumor effect [158]. Given the role of PI3K/AKT in priming chromatin for transcriptional activation, inhibiting its kinase activity can cause transcriptional repression of oncogenes, such as MYC [133].
