Monomeric GTPases, which belong to the Ras superfamily, are small proteins involved in many biological processes. The most studied families are Ras, Rho, Rab, Ran, Arf, and Miro, and recently, a new family named Big Ras GTPases was reported. As a general rule, the proteins of all families have five characteristic motifs (G1–G5), and some specific features for each family have been described. The main functions described for monomeric GTPases in fungi include morphogenesis, secondary metabolism, vesicle trafficking, and virulence.
- small GTPases
- Ras
- Rho
- Rab
- Ran
- Arf
- Miro
- BRG
- fungi
1. Overview
Small Ras GTPases (rat sarcoma guanine triphosphatases) are monomeric G proteins of low molecular weight (21 to 30 kDa) that are present in all eukaryotes and that participate in many biological processes. They act as molecular switches, alternating between an active GTP-bound state and an inactive GDP-bound state [1]. This regulation is carried out through the activity of guanine-nucleotide exchange factors (GEFs) and GTPase activating proteins (GAPs) that catalyze the replacement of GDP by GTP and promote the inactive GDP-bound form, respectively (Figure 1) [2,3,4][2][3][4].
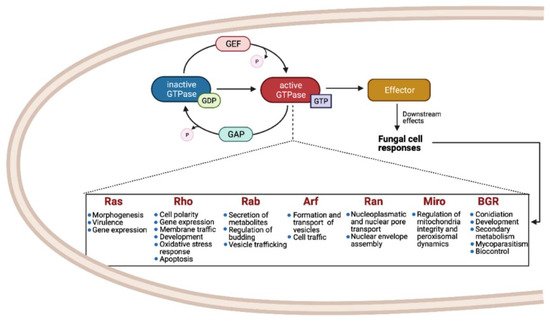
These proteins have a conserved GTP/GDP core and a hydrolysis domain bearing five characteristic and highly conserved motifs: G1 (Walker A/P-loop; GxxxxG K[S/T]) implicated in the binding of β- and γ-phosphate groups of the nucleotide; G2 (Switch I; x[T/S]x) that binds Mg2+, essential for GTP hydrolysis; G3 (Walker B/Switch II; DxxG) that interacts with the nucleotide γ-phosphate and Mg2; G4 ([N/T]KxD) that binds directly to the nucleotide at K and D sites; and G5 (S/CAK/L/T), involved in guanine base recognition [5,6][5][6]. The switch I and II regions are flexible segments that sense the nucleotide state and allow the small GTPases to interact with downstream effectors [7,8][7][8].
Members of the Small Ras-GTPases superfamily take part in cell regulation, growth, morphogenesis, cell division, and virulence and are known as master regulators of intracellular bidirectional vesicle transport [1,9][1][9]. They are classified according to their amino acid sequence and their biochemical properties.
As shown in Figure 1, the most common families and their main functions in fungi are Ras (rat sarcoma), associated with morphogenesis and virulence of fungi with different lifestyles, such as pathogens, saprobes, entomopathogens, and mutualists; Rho (Ras homologue) also known as Cdc42/Rho-GTPases, involved in the regulation of cell polarity, gene expression, and membrane traffic; Rab (Ras homologue from brain) that participates in the secretion of metabolites and lytic enzymes [10]; Arf (ADP ribosylation factors) participate in membrane/protein trafficking to the plasma membrane [9]; Ran (Ras-like nuclear proteins) play pivotal roles in nucleocytoplasmic and nuclear pore transport, synthesis of RNA, and as a checkpoint of the cell cycle [11]; and Miro (mitochondrial Rho-GTPases), also known as atypical Rho GTPases, serves critical roles in mitochondria morphology, inheritance, and homeostasis [12]. Other less studied families have been reported, such as Era-like (Escherichia coli Ras-like protein), involved in ribosomal assembly [13], and Spg1/Tem1 (Septum promoting) that participate in ring constriction, septation, and cell division in Schizosaccharomyces pombe [14]. Recently, a new fungal family of Ras GTPases was reported, the Big Ras GTPases (BRG), whose main difference is their size, 3- to 4-times bigger compared with the classical small GTPases as well as with their poorly conserved G4 motif. The only member characterized until now, TBRG-1 of the plant mutualistic fungus Trichoderma virens, is involved in conidiation, development, secondary metabolism, mycoparasitism, and biocontrol [15]. Proteins from the different families crosstalk with each other and with members of other signal transduction pathways, which together comprise a very large and complex network [9].
2. Classification and Phylogenetic Conservation of Small Ras GTPases in Fungi
Fungi are eukaryotic organisms that are highly relevant ecologically. They differ from plants due to their heterotrophic nature, and from animals because they possess cell walls. Their closest kingdom is the Animalia, from which they diverged nearly 1.3 billion years ago [16,17][16][17]. One of their major features is osmotrophy, a way of feeding and nourishment that involves the displacement of degraded organic compounds by osmosis. Fungi secrete lytic enzymes to breakdown the molecules and substrates so they can be passed through their cell walls for feeding [18]. This mechanism is used by many fungal species to compete among themselves or with other microorganisms. Fungi have evolved several strategies to degrade hard-to-digest substrates, which are also useful for establishing a relationship with plants. Moreover, they secrete a plethora of secondary metabolites [19]. The small Ras GTPases play important roles in several of these biological processes in fungi.
During the last two decades, genomic tools have greatly helped to study the phylogenetic relationships between different species of organisms, as well as to analyze complete gene families. With this purpose, James and coworkers [17] analyzed the current state of the fungal tree of life and reported that 224 orders of fungi are distributed into 12 phyla. The Ascomycota phylum contains the greatest number of species, followed by Basidiomycota, which together with Entorrhizomycota form the subkingdom Dikarya. The sister of this subkingdom is Mucoromycota, which contains mostly plant-associated taxa. Other important phyla are Zoopagomycota, related with parasitic and predatory behaviors, and sometimes saprobic; Blastocladiomycota, which is unique, in having an alteration of haploid and diploid generations and whose members have uniflagellated zoospores; and Chytridiomycota, which is composed of Aphelidiomycota, Cryptomycota/Rozellomycota, and Microsporidia, a group endoparasitic zoosporic fungi placed at the basal branches of the fungal kingdom [17].
In this work, we first focus on analyzing the members of the small Ras GTPase superfamily using genomic data. To find which families of Ras GTPases are present in the fungal kingdom and to search for the relationship between their phylogenetic data and their lifestyles, we first selected 56 genomes of different species distributed in eight different phyla (Figure 2, Table S1). All protein sequences were obtained from “The Fungal Genomics Resource” (MycoCosm. Available online: https://mycocosm.jgi.doe.gov/mycocosm/home, accessed on 14 March 2021) from the Joint Genome Institute (JGI) [20], using three search criteria: (1) PF00071, the Pfam domain key to Ras family, which displays results for the Ras, Rho, Rab, and Ran families; (2) PF00025, ADP ribosylation factor for the members of the Arf family; and (3) PF08477, the Ras complex, Roc, domain of DAPkinase, to find the members of the Miro family. To find the putative Big Ras GTPases, we performed a BLASTP using the sequence of TBRG-1 with each genome analyzed. As shown in Figure 2, the Ras, Rho, Rab, Ran, Arf, Miro, and BRG families are present in fungi, with the Arf family being the most abundant, whereas the Ran family contains the lowest number of members among all species. These patterns of distribution have also been observed in organisms of other kingdoms, such as Arabidopsis thaliana, Oryza sativa, Solanum lycopersicum, Zea mays, Lotus japonicas, Medicago truncatula, Phaseolus vulgaris, and Glycine max from plants; and Homo sapiens and Drosophila melanogaster from animals [21,22,23][21][22][23]. Intriguingly, only few species showed a hit with BRG, Ascomycota being the most represented phylum (magenta labels, Figure 2).
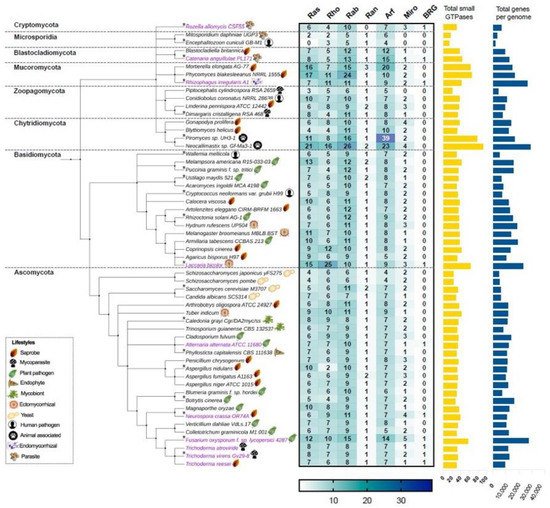
Figure 2. The Ras, Rho, Rab, Ran, Arf, Miro, and BRG families of the small Ras GTPases superfamily are present in the fungal kingdom. The taxonomic tree was generated using phyloT based on the NCBI taxonomy. Each phylum is indicated in its corresponding cluster. The numbers of members of each family are indicated in the heat-map table, which were obtained by searching in MycoCosm of JGI using PF00071, PF00025, and PF08477 keys as search criteria. The total number of small GTPases for each species are indicated in the yellow bars. The total number of genes of each genome were also obtained in MycoCosm and are indicated in blue bars. A representative lifestyle for each strain is indicated with icons. Those strains that contain putative BRGs in their genomes are indicated in magenta. The asterisks indicate the 20 species that were selected for further analysis.
With regard to the selected ascomycetes, Fusarium oxysporum has the greatest number of Ras GTPases with 57 members compared with the remaining species, whereas the fission yeasts Schizosaccharomyces japonicus yFS275 and Schizosaccharomyces pombe have only 23 elements each, being the less represented species. In basidiomycetes, Laccaria bicolor showed 63 members of Ras GTPases and Wallemia mellicola 30, where the first one bears the higher number of Rho GTPases of the analyzed fungi (Figure 2). In the phylum Chytridiomycota, we found the species with the highest number of Ras GTPases. Neocallimastix sp. Gf-Ma3-1 bears the highest number with 92 members, being the species with the major number of Ras and Rab families, whereas Piromyces sp. UH3-1 has 77 members, with a huge number of Arf proteins (Figure 2). Intriguingly, both species are animal-associated fungi. In Mucoromycota, we found two other species well represented: Phycomyces blakesleeanus NRRL1555 and Mortierella elongata AG-77, with 65 and 63 elements each, respectively. In contrast, Encephalitozoon cuniculi GB-M1 and Mtosporidium daphniae UGP3 that belong to the Microsporidia phylum bear 13 and 16 members, respectively, resulting in their being the species with the smallest number of Ras GTPases. Most of the remaining analyzed species ranged between 30 and 40 components of Ras GTPases superfamily, distributed in six families. These distributions in Ras GTPases members correlates well with the total number of genes in each genome (Figure 2, Table S1). Kelkar and Ochman point out that the wide variation in fungal genomes’ sizes could be related to their lifestyles [24]. In Laccaria bicolor, which establishes a mutualistic association with plant roots, the so-called ectomycorrhiza (ECM), it has been proposed that its large genome size resulted from an expansion of gene families’ sizes. This expansion includes protein kinases and Ras GTPases, which overcame a large number of young duplicates or paralogs after the separation from their sister Coprinopsis cinerea (saprobe, Figure 2) [25]. Additionally, during this evolution process, in L. bicolor members of Ras GTPases have retained, gained, or lost motifs compared with their ancestors, which resulted in the formation of pseudogenes or proteins with new functions. In this genome expansion, those genes related to their symbiotic lifestyle are present [25]. In contrast, members of microsporidia are intercellular parasites of agricultural and medical importance whose genomes are characterized by their reduction in the number of genes. This genome compaction is due to their reduced intergenic spaces and the shortness of most putative proteins relative to their eukaryote orthologues. Furthermore, they lack mitochondria and peroxysomes [26]. In agreement with this, E. cuniculi GB-M1 and M. daphniae UGP3 here analyzed barely contain 1996 and 3330 genes in their genomes, respectively, and lack Miro Ras GTPases, the mitochondrial Rho atypical proteins (Figure 2, Table S1).
To investigate the phylogenetic relationships among the seven families of the small Ras GTPases in fungi, we selected 20 species distributed in the eight phyla (denoted by an asterisk, Figure 2), and a single gene of each family was randomly selected to construct a phylogenetic tree (Figure 3). The members of distinct families were grouped, except for putative BRGs, such as those of Catenaria anguillulae PL171 and Rozella allomycis CSF55, and Rhizophagus irregularis A1, which were grouped with Rho and Ran families, respectively. Three proteins annotated in JGI as Ras were grouped with the Rab family (gray labels, Figure 3). As we expected, the subfamilies of each family were also grouped in distinct clades. As shown in Figure 3, the Rab18, Rab1/YPT1, SEC4, Rab5/YPT51, Rab6/YPT6/Ryh1, Rab11/YPT3, Rab2, and Rab4 all from the Rab family are clustered. For the Arf family, we distinguished Arf1, Arl3, Arf6, Arl2, Arl1, and Arf6 to be clustered, with a few exceptions. Similar results were reported by Schmoll et al. (2016) for an analysis of Ras GTPases families using three species of Trichoderma (T. virens, T. atroviride, and T. reesei), where the Ras and Rho families derived from the same branch; likewise, both Ran and Arf families derived from the same branch [1]. However, they reported two main branches for these Trichoderma species, whereas we found four. This could be explained by the fact that we used a greater number of species with different evolutionary processes. Overall, the proteins of the same phylum were clustered together, being more evident for Ascomycota and Basidiomycota because more species of these phyla were included.
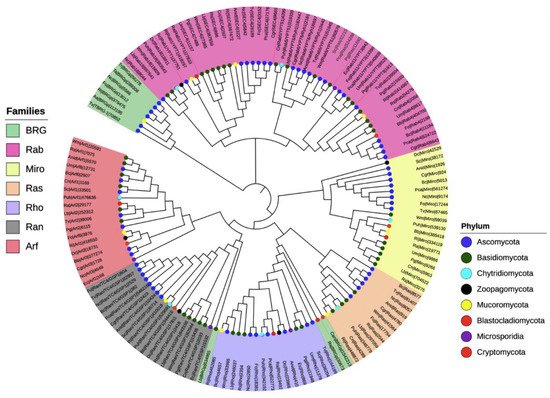
Figure 3. Phylogenetic tree of the seven families of the small Ras GTPases in fungi. The evolutionary history was inferred using the neighbor-joining method [27] using 144 proteins. The bootstrap test was used with 1500 replicates [28]. The evolutionary distances were computed using the Poisson correction method. Evolutionary analyses were conducted in MEGA7 [29].
(References would be added automatically after the entry is online)
References
- Schmoll, M.; Dattenböck, C.; Carreras-Villaseñor, N.; Mendoza-Mendoza, A.; Tisch, D.; Alemán, M.I.; Baker, S.E.; Brown, C.; Cervantes-Badillo, M.G.; Cetz-Chel, J.; et al. The genomes of three uneven siblings: Footprints of the lifestyles of three Trichoderma species. Microbiol. Mol. Biol. Rev. 2016, 80, 205–327.
- Barrett, T.; Xiao, B.; Dodson, E.J.; Dodson, G.; Ludbrook, S.B.; Nurmahomed, K.; Gamblin, S.J.; Musacchio, A.; Smerdon, S.J.; Eccleston, J.F. The structure of the GTPase-activating domain from p50rhoGAP. Nature 1997, 385, 458.
- Wennerberg, K.; Rossman, K.L.; Der, C.J. The Ras superfamily at a glance. J. Cell Sci. 2005, 118, 843–846.
- van Dam, T.J.P.; Bos, J.; Snel, B. Evolution of the Ras-like small GTPases and their regulators. Small GTPases 2011, 2, 4–16.
- Francis, S.M.; Gas, M.-E.; Daugeron, M.-C.; Bravo, J.; Seraphin, B. Rbg1–Tma46 dimer structure reveals new functional domains and their role in polysome recruitment. Nucleic Acids Res. 2012, 40, 11100–11114.
- Bourne, H.R.; Sanders, D.A.; McCormick, F. The GTPase superfamily: Conserved structure and molecular mechanism. Nature 1991, 349, 117–127.
- Macara, I.G.; Lounsbury, K.M.; Richards, S.A.; McKiernan, C.; Bar-Sagi, D. The Ras superfamily of GTPases 1. FASEB J. 1996, 10, 625–630.
- Wittinghofer, A.; Vetter, I.R. Structure-function relationships of the G domain, a canonical switch motif. Annu. Rev. Biochem. 2011, 80, 943–971.
- Just, W.W.; Peränen, J. Small GTPases in peroxisome dynamics. Biochim. Biophys. Acta (BBA) Mol. Cell Res. 2016, 1863, 1006–1013.
- Pereira-Leal, J.B.; Seabra, M.C. Evolution of the Rab family of small GTP-binding proteins. J. Mol. Biol. 2001, 313, 889–901.
- Rojas, A.M.; Fuentes, G.; Rausell, A.; Valencia, A. The Ras protein superfamily: Evolutionary tree and role of conserved amino acids. J. Cell Biol. 2012, 196, 189–201.
- Fransson, Å.; Ruusala, A.; Aspenström, P. Atypical Rho GTPases have roles in mitochondrial homeostasis and apoptosis. J. Biol. Chem. 2003, 278, 6495–6502.
- Takai, Y.; Sasaki, T.; Matozaki, T. Small GTP-binding proteins. Physiol. Rev. 2001, 81, 153–208.
- Schmidt, S.; Sohrmann, M.; Hofmann, K.; Woollard, A.; Simanis, V. The Spg1p GTPase is an essential, dosage-dependent inducer of septum formation in Schizosaccharomyces pombe. Genes Dev. 1997, 11, 1519–1534.
- Dautt-Castro, M.; Estrada-Rivera, M.; Olguin-Martínez, I.; del Rocha-Medina, M.C.; Islas-Osuna, M.A.; Casas-Flores, S. TBRG-1 a Ras-like protein in Trichoderma virens involved in conidiation, development, secondary metabolism, mycoparasitism, and biocontrol unveils a new family of Ras-GTPases. Fungal Genet. Biol. 2020, 136, 103292.
- Berbee, M.L.; James, T.Y.; Strullu-Derrien, C. Early diverging fungi: Diversity and impact at the dawn of terrestrial life. Annu. Rev. Microbiol. 2017, 71, 41–60.
- James, T.Y.; Stajich, J.E.; Hittinger, C.T.; Rokas, A. Toward a fully resolved fungal tree of life. Annu. Rev. Microbiol. 2020, 74.
- Richards, T.A.; Talbot, N.J. Osmotrophy. Curr. Biol. 2018, 28, R1179–R1180.
- Rokas, A.; Mead, M.E.; Steenwyk, J.L.; Raja, H.A.; Oberlies, N.H. Biosynthetic gene clusters and the evolution of fungal chemodiversity. Nat. Prod. Rep. 2020, 37, 868–878.
- Grigoriev, I.V.; Nikitin, R.; Haridas, S.; Kuo, A.; Ohm, R.; Otillar, R.; Riley, R.; Salamov, A.; Zhao, X.; Korzeniewski, F. MycoCosm portal: Gearing up for 1000 fungal genomes. Nucleic Acids Res. 2014, 42, D699–D704.
- Vernoud, V.; Horton, A.C.; Yang, Z.; Nielsen, E. Analysis of the small GTPase gene superfamily of Arabidopsis. Plant Physiol. 2003, 131, 1191–1208.
- Flores, A.C.; Via, V.D.; Savy, V.; Villagra, U.M.; Zanetti, M.E.; Blanco, F. Comparative phylogenetic and expression analysis of small GTPases families in legume and non-legume plants. Plant Signal. Behav. 2018, 13, e1432956.
- Jiang, S.-Y.; Ramachandran, S. Comparative and evolutionary analysis of genes encoding small GTPases and their activating proteins in eukaryotic genomes. Physiol. Genom. 2006, 24, 235–251.
- Kelkar, Y.D.; Ochman, H. Causes and consequences of genome expansion in fungi. Genome Biol. Evol. 2012, 4, 13–23.
- Rajashekar, B.; Kohler, A.; Johansson, T.; Martin, F.; Tunlid, A.; Ahrén, D. Expansion of signal pathways in the ectomycorrhizal fungus Laccaria bicolor—evolution of nucleotide sequences and expression patterns in families of protein kinases and RAS small GTPases. New Phytol. 2009, 183, 365–379.
- Katinka, M.D.; Duprat, S.; Cornillot, E.; Méténier, G.; Thomarat, F.; Prensier, G.; Barbe, V.; Peyretaillade, E.; Brottier, P.; Wincker, P. Genome sequence and gene compaction of the eukaryote parasite Encephalitozoon cuniculi. Nature 2001, 414, 450–453.
- Saitou, N.; Nei, M. The neighbor-joining method: A new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 1987, 4, 406–425.
- Felsenstein, J. Confidence limits on phylogenies: An approach using the bootstrap. Evolution 1985, 39, 783–791.
- Kumar, S.; Stecher, G.; Tamura, K. MEGA7: Molecular Evolutionary Genetics Analysis Version 7.0 for Bigger Datasets. Mol. Biol. Evol. 2016, 33, 1870–1874.
