Periodontitis is a chronic complex inflammatory disease associated with a destructive host immune response to microbial dysbiosis, leading to irreversible loss of tooth-supporting tissues.
- circular RNAs
- epigenetics
- pathogenesis
- biomarkers
- periodontal regeneration
- osteogenic differentiation
1. Periodontal Disease and Regeneration
Periodontal Disease
Periodontal diseases include gingivitis and periodontitis, both of which are widespread chronic inflammatory diseases [1,2][1][2]. Periodontitis occurs as a destructive host immune response to the accumulation of a bacterial biofilm (dental plaque) on the root surfaces of teeth. Inflammatory changes lead to progressive and episodic bursts of destruction of the supporting tissues around the teeth, which can lead to eventual tooth loss [3]. The global burden of periodontal disease increased by 57.3% from 1990 to 2010 [4]. Severe forms of periodontitis affect around 11% of adults across the globe [4,5][4][5]. At the global level, aggressive forms of periodontitis (also called ‘juvenile periodontitis’) affect up to 1.6% of people who are younger than 30 years old, including children and adolescents [6].
A healthy periodontium consists of soft tissues (the gingiva and periodontal ligament) and hard tissues (cementum and alveolar bone) (Figure 1a). The accumulation of a complex microbial biofilm (dental plaque) at the interface of the tooth and the adjacent gingiva results in inflammation (gingivitis). Gingivitis is reversible once the plaque biofilm is disrupted and removed. In contrast, in susceptible patients, dental plaque induces irreversible inflammation of the underlying periodontal tissues, resulting in loss of attachment and resorption of bone. When inflammation extends deep into tissues, it results in extensive destruction of both soft and hard tissues, and this might ultimately cause loss of the affected tooth (Figure 1b,c) [7]. The likelihood of this happening is determined by both inherited factors such as cytokine gene polymorphisms and other immune response genes and inherited factors, the presence of conditions that modify the host’s response (such as diabetes mellitus and smoking), and local factors that predispose to the stagnation and maturation of the dental plaque biofilm (such as oral appliances and dentures) [8,9,10,11,12,13,14][8][9][10][11][12][13][14].
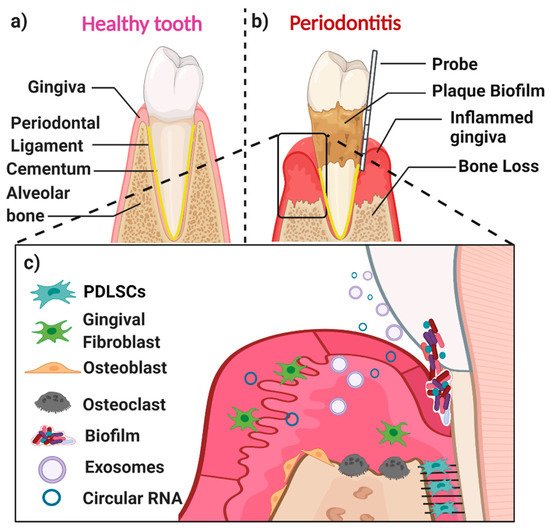
The main cell populations found within a healthy or diseased periodontium are periodontal ligament (stem) cells (PDLSCs), gingival fibroblasts (GFs), osteoblasts (OBs), osteoclasts, and various immune cells [15] (Figure 1c). An important biofluid in the context of the periodontium is gingival crevicular fluid (GCF), which is derived from serum. It flows passively from the gingival crevice, at the interface between the tooth and the adjacent gingiva. The composition of GCF alters according to the disease status of a particular periodontal site. The flow is greater when the site is inflamed. GCF can contain neutrophils as well as various cell-derived biological molecules, including small extracellular vesicles [16,17,18,19][16][17][18][19] and circular RNAs (circRNAs). These small molecules are also present in the periodontal tissues, where they play an essential role in periodontal tissue homeostasis, repair, and pathogenesis.
Susceptibility to periodontitis is influenced by both genetic and epigenetic factors, as well as by factors that modify the host immune response, such as smoking and uncontrolled diabetes mellitus [8,11,20][8][11][20]. Epigenetics is defined as a stably heritable phenotype resulting from chemical changes in a chromosome, without causing any changes in the DNA sequence, including DNA methylation, histone modification, and non-coding RNAs (ncRNA). Epigenetic-associated ncRNAs can be divided into two main groups—small ncRNAs (<30 nts) and long ncRNA (>200 nts). The potential role of epigenetics-related ncRNAs in diagnosing periodontitis was investigated previously in PDL and gingiva tissues [21,22,23,24][21][22][23][24];
2. circRNA Expression in Periodontal Tissues
There is an emerging understanding of the potential role of circRNA in periodontitis, and in particular, disease pathogenesis. There have been three studies on their presence in PDL tissues [37,38,39][25][26][27] and two studies in gingival tissues [40,41][28][29] taken from periodontitis patients (Table 1).
Table 1. Expression of circRNAs in PDL (and cells) and gingival tissues.
| Reference | circRNA Name (Gene Name), Genome Location and Length | circRNA Target or Pathway | CircRNA Detection Method |
Cell (Tissue) Details | Key Findings |
|---|---|---|---|---|---|
| Yu et al. 2021 [37][25] | circMAP3K11 (MAP3K11) | miR-511 | RT-qPCR10 | PDL tissues from 10 healthy and 20 periodontitis cases. hPDLSCs from healthy donors. Donor age—unclear. Passage 3. |
Higher expression levels of circMAP3K11 and TLR4, and lower expression levels of miR-511-3p were found in periodontitis affected PDL tissues, compared to healthy controls. circMAP3K11 enhanced hPDLSCs proliferation, migration and osteogenic differentiation, and reduced the apoptosis of hPDLSCs in vitro through a circMAP3K11/ miR-511-3p/TLR4 axis. In vivo—silencing circMAP3K11 can prevent periodontitis development in mice, with decreased cell proliferation and increased apoptosis. |
| Zheng et al. 2021. [38][26] | hsa_circ_0003489 (circCDK8) chr13:26974589-26975761; 1172 bp | mTOR signalling pathway | RT-qPCR | PDL tissues from 6 healthy and 6 mild/moderate chronic periodontitis patients hPDLSCs; Healthy PDL tissues (third molars); Passage 3 to 5. |
CircCDK8 and HIF-1α were increased in PDL tissues from periodontitis patients. Overexpression of CircCDK8 decreased osteogenic differentiation of hPDLSCs through the mTOR pathway under hypoxia. |
| Wang et al. 2019. [39][27] | hsa_circ_0001946; CDR1as (or CiRS-7) chrX:139865339-139866824; 1485 bp |
miR-7 ERK/MAPK signal pathway |
RT-qPCR | PDL tissue from 10 periodontitis and 11 healthy cases. hPDLSCs from healthy teeth and chronic periodontitis tissue; passage 3. Donor age 30–40 yrs. |
circCDR1as was significantly downregulated in PDL tissues from periodontitis patients. circRNA CDR1as inhibited hPDLSCs proliferation through the miR-7 and ERK/MAPK pathway |
| Wang et al. 2021. [40][28] | circ_0081572 (GRHL2) chr8:102564942-102571040; 6098 bp |
miR-378h | RT-qPCR | Gingival tissues from 21 healthy and 21 periodontitis cases, Human periodontal ligament cells (hPDLCs). Unclear donor age and cell passage number. |
Circ_0081572 was downregulated in the gingival tissues of periodontitis, compared to healthy gingival tissues. Overexpression of circ_0081572 could alleviate LPS-induced PDLCs injury via circ_0081572/miR-378h/RORA axis. |
| Li et al. 2019. [41][29] | hsa_circ_0062491 chr22:23063339-23180200; 116861 bp |
miR-584 | RNA sequencing RT-qPCR |
Gingival tissues from 4 healthy and 4 chronic periodontitis cases. THP1 cells treated with P. gingivalis. |
1, 304 circRNAs were significantly differentially expressed in the gingival tissues of periodontitis patients (n = 4). Decreased circ_0062491 and increased circ_0095812 found in periodontitis-gingival tissues compared to healthy tissues (n = 30) using RT-qPCR. Circ_0062491 function as a miR-584 sponge in THP1 cells. |
Abbreviations: MAP3K11: Mitogen-activated protein kinase kinase kinase 11; CDK8: Cyclin-Dependent Kinase 8; CDR1as: Cerebellar degeneration-related protein 1 antisense RNA;GRHL2: grainyhead like transcription factor 2; TLR4: Toll-like receptor 4; HIF-1α: Hypoxia-inducible factor 1-alpha; mTOR: The mechanistic target of rapamycin; ERK: extracellular-signal-regulated kinase; RORA: RAR-related orphan receptor alpha; and THP1: human acute monocytic leukemia cell line.
A very recent study by Yu et al. [37][25] demonstrated that circMAP3K11 (also named hsa_circ_002284) was significantly increased in inflamed PDL tissues from periodontitis (n = 20), as compared to healthy PDL tissues (n = 10), using RT-qPCR. Periodontitis patients were selected based on the following criteria—(1) redness and swelling of the gingiva or bleeding on probing; (2) probing depth > 3 mm and attachment loss > 1 mm; and (3) radiographic evidence of horizontal or vertical resorption of alveolar bone. However, these criteria represent a low threshold for defining periodontitis, and the latest stage and grade guidelines for classifying periodontitis were not used. Furthermore, circMAP3K11 enhanced hPDLSCs proliferation and migration and reduced the apoptosis of hPDLSCs in vitro via the circMAP3K11/miR-511-3p/TLR4 axis. Overexpression of circMAP3K11 led to increased proliferation, migration, and osteogenic protein expression of Runx2, OSX, ATF4, and BSP in hPDLSCs.The dual-luciferase reporter assay demonstrated that miR-511-3p directly targeted circMAP3K11, and that toll-like receptor 4 (TLR4) is a direct target of miR-511-3p. Additionally, a mouse periodontitis disease model was generated in 24 female 8-week-old C57BL/6 J mice. After oral injection of hPDLSCs transfected with si-circMAP3K11 (1 × 104 cells/mice) for 48 h, the si-circMAP3K11 group exhibited the highest apoptosis and decreased proliferating cells; indicating that circMAP3K11 promoted the proliferation and inhibited the apoptosis of hPDLSCs by acting as an miR-511-3p sponge in vivo.
Zheng et al. [38][26] revealed that hsa_circ_0003489 was significantly increased in PDL tissues from patients with periodontitis (n = 6) compared to a healthy control group. Hsa_circ_0003489 is located at the gene for cyclin-dependent kinase 8 (CDK8) and is also known as circCDK8. Six samples of either healthy or inflamed PDL tissues were collected. The inflamed samples were sourced from patients with mild or moderate chronic periodontitis. The healthy samples were collected from teeth extracted for orthodontic treatment or from erupted third molars. Using RT-qPCR, it was found that circCDK8 was expressed at more than two-fold higher levels in periodontitis compared with control tissues. circCDK8 was shown to inhibit the osteogenic differentiation of hPDLSCs (passage 3–5) under hypoxic conditions. Overexpression of circCDK8 leads to autophagy and apoptosis of PDLSCs via the mTOR signalling pathway. On the other hand, silencing of circCDK8 using small interfering RNA (si-circCDK8) enhances osteogenic differentiation in hPDLSCs through altered mTOR signalling [38][26].
Wang et al. [39][27] found that circRNA of cerebellar degeneration-related protein 1 transcript (circCDR1as) was significantly downregulated in PDL tissues from periodontitis patients. In this particular study, human PDL tissues were obtained from teeth with chronic periodontitis that was extracted because of advanced disease (n = 10), or from healthy teeth removed for orthodontic purposes (n = 11). Once again, using RT-qPCR, it was found that the expression level of CDR1as was significantly lower in PDL tissues from sites with periodontitis as compared to normal tissues. circCDR1as was also downregulated in lipopolysaccharide (LPS)-treated hPDLSCs. Knockdown of CDR1as (using siRNA-CDR1as) enhanced LPS-induced proliferative inhibition of hPDLSCs. In contrast, overexpression of CDR1as (by a pcDNA3.1-circ-mini-CDR1as plasmid) was found to promote the proliferation of hPDLSCs. Additionally, circCDR1as functions as a sponge for miR-7, and this is relevant to the extracellular-signal-regulated extracellular-signal-regulated kinase (ERK) or MAPK signalling pathway during the proliferation of hPDLSCs.
A recent study by Wang et al. [40][28] used RT-qPCR to show that circ_0081572 was significantly decreased in periodontitis-derived gingival tissues (n = 21) compared to healthy gingival tissues (n = 21). circ_0081572 was mainly distributed in the cytoplasm of human periodontal ligament cells (hPDLCs). Overexpression of circ_0081572 reversed the effects of lipopolysaccharide (LPS), which caused decreased viability and stimulated apoptosis and caspase 3 activity of PDLCs, by targeting miR-378h and retinoid acid-related orphan receptor A (RORA). This suggests that circ_0081572 inhibits the progression of periodontitis through regulation of the miR-378h/RORA axis.
Using RNA sequencing, Li et al. [41][29] found that a total of some 1,304 circRNAs were significantly expressed in the gingival tissues of periodontitis patients (n = 4). The authors selected six altered circRNAs and validated their presence using RT-qPCR. In total, in periodontitis gingival tissues, the levels of four circRNAs were increased (circ_0099630, circ_0138960, circ_0138959, and circ_0107783), while the level of the other two decreased (circ_0062490 and circ_0085289) when compared to healthy controls. In periodontitis tissues, circ_0062491 was significantly decreased, while circ_0095812 was significantly increased (n = 30) when compared to control tissues (n = 30).
In summary, circMAP3K11, circCDK8, circCDR1as, circ_0081572, circ_0062491, and circ_0095812 are potentially associated with periodontitis; however, further studies are required to confirm these findings. The literature on the role of circRNAs in disease pathogenesis is limited, and this is an area that clearly requires further investigation.
(References would be added automatically after the entry is online)
References
- Bartold, P.M.; Van Dyke, T.E. Periodontitis: A host-mediated disruption of microbial homeostasis. Unlearning learned concepts. Periodontology 2000 2013, 62, 203–217.
- Billings, M.; Holtfreter, B.; Papapanou, P.N.; Mitnik, G.L.; Kocher, T.; Dye, B.A. Age-dependent distribution of periodontitis in two countries: Findings from NHANES 2009 to 2014 and SHIP-TREND 2008 to 2012. J. Clin. Periodontol. 2018, 45, S130–S148.
- Kinane, D.F.; Bartold, P.M. Clinical relevance of the host responses of periodontitis. Periodontology 2000 2007, 43, 278–293.
- Kassebaum, N.J.; Bernabé, E.; Dahiya, M.; Bhandari, B.; Murray, C.J.L.; Marcenes, W. Global Burden of Severe Periodontitis in 1990–2010: A Systematic Review and Meta-Regression; SAGE Publications: Los Angeles, CA, USA, 2014; Volume 93, pp. 1045–1053.
- Chapple, I.L.C.; Van der Weijden, F.; Doerfer, C.; Herrera, D.; Shapira, L.; Polak, D.; Madianos, P.; Louropoulou, A.; Machtei, E.; Donos, N.; et al. Primary prevention of periodontitis: Managing gingivitis. J. Clin. Periodontol. 2015, 42, S71–S76.
- Bouziane, A.; Hamdoun, R.; Abouqal, R.; Ennibi, O. Global prevalence of aggressive periodontitis: A systematic review and meta-analysis. J. Clin. Periodontol. 2020, 47, 406–428.
- Hienz, S.A.; Paliwal, S.; Ivanovski, S. Mechanisms of bone resorption in periodontitis. J. Immunol. Res. 2015.
- Bartold, P.M. Lifestyle and periodontitis: The emergence of personalized periodontics. Periodontology 2000 2018, 78, 7–11.
- Chapple, I.L.C.; Mealey, B.L.; Van Dyke, T.E.; Bartold, P.M.; Dommisch, H.; Eickholz, P.; Geisinger, M.L.; Genco, R.J.; Glogauer, M.; Goldstein, M.; et al. Periodontal health and gingival diseases and conditions on an intact and a reduced periodontium: Consensus report of workgroup 1 of the 2017 World Workshop on the Classification of Periodontal and Peri-Implant Diseases and Conditions. J. Clin. Periodontol. 2018, 45 (Suppl. 20), S68–S77.
- Lang, N.P.; Kiel, R.A.; Anderhalden, K. Clinical and microbiological effects of subgingival restorations with overhanging or clinically perfect margins. J. Clin. Periodontol. 1983, 10, 563–578.
- Meyle, J.; Chapple, I. Molecular aspects of the pathogenesis of periodontitis. Periodontology 2000 2015, 69, 7–17.
- Mizutani, S.; Ekuni, D.; Tomofuji, T.; Azuma, T.; Kataoka, K.; Yamane, M.; Iwasaki, Y.; Morita, M. Relationship between xerostomia and gingival condition in young adults. J. Periodontal Res. 2015, 50, 74–79.
- Murakami, S.; Mealey, B.L.; Mariotti, A.; Chapple, I.L.C. Dental plaque-induced gingival conditions. J. Periodontol. 2018, 89 (Suppl. 1), S17–S27.
- Offenbacher, S.; Barros, S.P.; Beck, J.D. Rethinking periodontal inflammation. J. Periodontol. 2008, 79, 1577–1584.
- Ivanovski, S.; Gronthos, S.; Shi, S.; Bartold, P.M. Stem cells in the periodontal ligament. Oral Dis. 2006, 12, 358–363.
- Han, P.; Bartold, P.M.; Salomon, C.; Ivanovski, S. Salivary outer membrane vesicles and DNA methylation of small extracellular vesicles as biomarkers for periodontal status–a pilot study. Int. J. Mol. Sci. 2021, 22, 2423.
- Han, P.; Lai, A.; Salomon, C.; Ivanovski, S. Detection of salivary small extracellular vesicles associated inflammatory cytokines gene methylation in gingivitis. Int. J. Mol. Sci. 2020, 21, 5273.
- Han, P.; Ivanovski, S. Saliva-Friend and Foe in the COVID-19 Outbreak. Diagnostics 2020, 10, 290.
- Han, P.; Bartold, P.M.; Salomon, C.; Ivanovski, S. Salivary small extracellular vesicles associated miRNAs in periodontal status—A pilot study. Int. J. Mol. Sci. 2020, 21, 2809.
- Bartold, P.M.; Van Dyke, T.E. An appraisal of the role of specific bacteria in the initial pathogenesis of periodontitis. J. Clin. Periodontol. 2019, 46, 6–11.
- Jurdziński, K.T.; Potempa, J.; Grabiec, A.M. Epigenetic regulation of inflammation in periodontitis: Cellular mechanisms and therapeutic potential. Clin. Epigenet. 2020, 12, 186.
- Benakanakere, M.R.; Finoti, L.; Palioto, D.B.; Teixeira, H.S.; Kinane, D.F. Epigenetics, inflammation, and periodontal disease. Curr. Oral Health Rep. 2019, 6, 37–46.
- Larsson, L.; Castilho, R.M.; Giannobile, W.V. Epigenetics and its role in periodontal diseases: A state-of-the-art review. J. Periodontol. 2015, 86, 556–568.
- Larsson, L. Current concepts of epigenetics and its role in periodontitis. Curr. Oral Health Rep. 2017, 4, 286–293.
- Yu, B.; Hu, J.; Li, Q.; Wang, F. CircMAP3K11 contributes to proliferation, apoptosis and migration of human periodontal ligament stem cells in inflammatory microenvironment by regulating TLR4 via miR-511 sponging. Front. Pharmacol. 2021, 12.
- Zheng, J.; Zhu, X.; He, Y.; Hou, S.; Liu, T.; Zhi, K.; Hou, T.; Gao, L. CircCDK8 regulates osteogenic differentiation and apoptosis of PDLSCs by inducing ER stress/autophagy during hypoxia. Ann. N. Y. Acad. Sci. 2020, 1485.
- Wang, F.; Chen, X.; Han, Y.; Xi, S.; Wu, G. circRNA CDR1as regulated the proliferation of human periodontal ligament stem cells under a lipopolysaccharide-induced inflammatory condition. Mediat. Inflamm. 2019, 2019, 1625381.
- Wang, J.; Du, C.; Xu, L. Circ_0081572 inhibits the progression of periodontitis through regulating the miR-378h/RORA axis. Arch. Oral Biol. 2021, 124, 105053.
- Li, J.; Xie, R. Circular RNA expression profile in gingival tissues identifies circ_0062491 and circ_0095812 as potential treatment targets. J. Cell. Biochem. 2019, 120, 14867–14874.
