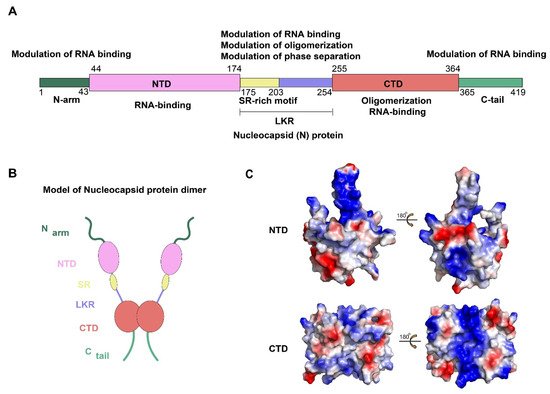
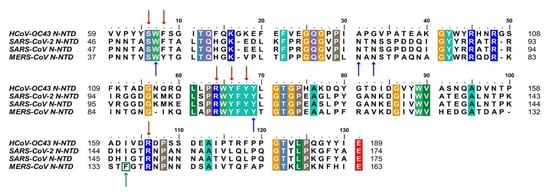
Figure 1. Sequences alignment of four CoVs N-NTD. Multiple sequence alignment of HCoV-OC43 (UniProtKB: P33469), SARS-CoV-2 (UniProtKB: P0DTC9), SARS-CoV (UniProtKB: P59595), MERS-CoV (UniProtKB: K9N4V7). The highly conserved residues were filled with colors. Red arrows indicate conserved RNA binding sites. Blue arrows and green arrow indicate conserved and mutant residues for the non-native interaction interface, respectively. HCoV-OC43, human coronavirus OC43; SARS-CoV-2, severe acute respiratory syndrome coronavirus 2; SARS-CoV, severe acute respiratory syndrome coronavirus; MERS-CoV, Middle East respiratory syndrome coronavirus. (The fill color selected in this figure legend is the default setting of the BioEdit software.).
In addition, blocking normal N protein oligomerization or triggering abnormal RNP formation is also an attractive inhibitory strategy. More recently, Lin et al. identified 5-benzyloxygramine (P3) is a novel inhibitor for MERS-CoV by virtual screening. This compound could mediate MERS-CoV N-NTD non-native dimerization and induce N protein aggregation. The structure-based study showed that P3 targets the non-native interface of N-NTD dimers and simultaneously interacts with the hydrophobic pockets in both N-NTD protomers. It was demonstrated that P3 was able to replace the vector-fusion residues of promoter 2 to occupy its binding cavity in promoter 1 under the legend free condition, which, in turn, stabilized the dimeric status by triggering massive hydrophobic interactions
[14][30][14,62]. By comparing the binding sites of P3 in the hydrophobic cavity, it was found that almost all of the residues of the N-NTD involved in the interactions are conserved, except F135 in MERS-CoV, which is replaced by I146 in SARS-CoV-2(). Although both residues are nonpolar amino acids, the effect on SARS-CoV-2 replication needs to be further verified. For other viruses, such as the human immunodeficiency virus and influenza virus, the researchers proposed a strategy to inhibit viral N protein oligomerization by developing competing peptides
[31][32][63,64]. For CoVs, it has been shown that the excessive peptide based on the C-terminal tail sequence can interfere with CTD oligomerization of HCoV-229E N protein and decrease the viral titer, providing a reference for relevant studies on SARS COV-2 N protein
[33][65]. Notably, the LLPS of N protein induced by viral genomic RNA is also a potential target
[34][35]. Slowing viral infection by increasing or decreasing the N protein LLPS is a strategy that could be considered. 1,6-hexanediol, lipoic acid, and aminoglycoside kanamycin, each of which potentially alters LLPS by a representative and distinct mechanism. In terms of SARS-CoV-2, further experiments showed that the formation or the size of condensates could be reduced after treatment with these small molecules
[35][34]. Meanwhile, high-throughput virtual screening is underway, several potential drug candidates have been proposed, and the next focus is on rigorous experimental validation, such as (−)-catechin gallate and (−)-gallocatechin gallate
[36][66] ().
Table 1. β-CoV inhibitors target N protein.
| Compounds |
Target Domain or Process |
Mechanism |
Reference |
| PJ34, N-(6-oxo-5,6-dihydrophenanthridin-2-yl) (N,N-dimethylamino) acetamide hydrochloride |
NTD |
Reduce RNA binding |
[14][37][14,67] |
| H3, 6-chloro-7-(2-morpholin-4-ylethylamino) quinoxaline-5,8-dione |
NTD |
Reduce RNA binding |
[29][61] |
| (−)-catechin gallate |
NTD |
Reduce RNA binding |
[36][66] |
| (−)-gallocatechin gallate |
NTD |
Reduce RNA binding |
[36][66] |
| P3, 5-benzyloxygr- amine |
CTD |
Induce abnormal dimerization |
[30][62] |
| 1,6-hexanediol |
LLPS |
prevent condensate formation |
[38][68] |
| Lipoic acid |
LLPS |
Reduce smaller condensate |
[38][68,6 |