Your browser does not fully support modern features. Please upgrade for a smoother experience.
Please note this is a comparison between Version 2 by Vivi Li and Version 1 by Shiming Peng.
FoxO1 is a conserved transcription factor involved in energy metabolism. It is tightly regulated by modifications on its mRNA and protein and responds to environmental nutrient signals. FoxO1 controls the transcription of downstream genes mediating metabolic regulation. Dysfunction of FoxO1 pathways results in several metabolic diseases, including diabetes, obesity, non-alcoholic fatty liver disease, and atherosclerosis.
- FoxO1
- metabolic disease
- drug discovery
1. Introduction of FoxO1
Forkhead Box O (FoxO) genes belong to a transcription factor (TF) family characterized by the presence of a conserved DNA-binding domain (the Forkhead box) in the N-terminal end of the protein [1,2,3][1][2][3] (Figure 1A). FOXOs bind the consensus DNA-binding element within the promoter of their target genes and regulate their transcription in response to external signal stimulations [4,5,6][4][5][6] (Figure 1B). There are four FoxO genes identified in mammals, including FoxO1, FoxO3a, FoxO4, and FoxO6 [7]. In the 1990s, the FoxO1 gene was first identified in studies of chromosomal translocations found in human tumors, including rhabdomyosarcomas and acute myeloid leukemias [1,3,8,9][1][3][8][9]. In addition to its important regulatory roles in oncogenesis [10], FoxO1 also transcriptionally mediates pathways behind many metabolic diseases, including gluconeogenesis, glycogenolysis, adipogenesis, thermogenesis, and feeding behavior [11[11][12][13][14][15],12,13,14,15], which is the focus of this review article.
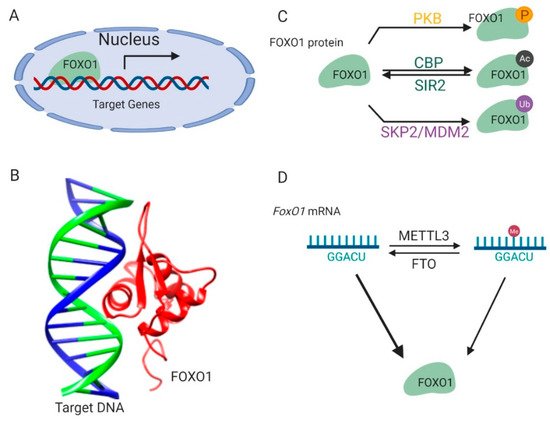
Figure 1. Regulation of FOXO1 transcription activity. (A) FOXO1 binds to the promoters of target genes and stimulates their transcription in nucleus. (B) The complex structure of the FOXO1 protein and its target-binding DNA (Protein Data Bank id: 3CO6). (C) The post-translational modifications of the FOXO1 protein. (D) Methylations on the FoxO1 mRNA regulate its expression. This figure was created with UCSF Chimera [29][16] and BioRender.com.
2. Regulation of FoxO1
2.1. Protein Modifications
FOXO1 protein is sophisticatedly and strictly regulated by modifications on its protein and mRNA, which ensures that transcription of its downstream target genes is tightly responsive to environmental signals [16][17] (Figure 1C). Kinase-mediated phosphorylation of the FOXO1 protein in response to insulin or growth factors is a form of regulation [17][18] (Figure 1C). Phosphoinositide 3-kinase/protein kinase B (PI3K/PKB) phosphorylates the FOXO1 protein at three conserved residues, including Threonine 24, Serine 256, and Serine 319. These modifications result in disrupted interactions between the FOXO1 protein and its target DNA and lead to the translocation of the FOXO1 protein from the nucleus to the cytoplasm, thus suppressing FOXO1-dependent transcription. The FOXO1 protein can also be phosphorylated by c-Jun N-terminal kinase (JNK) or macrophage-stimulating 1 (Mst1) [18,19][19][20]. This phosphorylation results in the import of the FOXO1 protein from the cytoplasm to the nucleus, thereby antagonizing the action of PI3K/PKB.
FOXO1 protein activity is also regulated by reversible acetylation modification on lysine residues (Figure 1C). They are acetylated by histone acetyltransferase cAMP-response element-binding protein (CREB)-binding protein (CBP) and deacetylated by NAD-dependent histone deacetylase silent information regulator 2 (Sir2) on conserved residues (Lys 242, Lys 245, and Lys 262) [20,21,22][21][22][23]. The positive charge of these lysine residues in FOXO1 contributes to DNA binding. Thus, acetylation at these residues attenuates the ability of FOXO1 to bind DNA and suppress transcription. Interestingly, it was also found that acetylation regulated the function of FOXO1 by influencing its sensitivity for phosphorylation.
The FOXO1 protein is polyubiquitinated by E3 ligases (Figure 1C) such as S-phase kinase-associated protein 2 (SKP2) and mouse double minute 2 homolog (MDM2), and targeted for protein degradation in human primary tumors and cancer cell lines [23,24,25][24][25][26]. The ubiquitination and proteasome degradation of the FOXO1 protein are essential in tumorigenesis and represent a viable target for cancer treatment.
2.2. mRNA Methylation
In addition to these post-translational modifications on protein, FoxO1 is also regulated by methylation modification on its mRNA [26,27,28][27][28][29] (Figure 1D). There are two specific adenosine sites in the coding sequence of the FoxO1 mRNA [26,27][27][28]. The adenosine (A) at these sites can be methylated to N6-Methyladenosine (m6A) by methyltransferase like 3 (METTL3) protein, and m6A can be demethylated to A by fat mass and obesity-associated protein (FTO). These reversible m6A modifications participate in regulating FoxO1 gene translation [28][29].
3. FoxO1-Regulating Mechanism Behind Diseases
3.1. Glucose Production in the Liver
FoxO1 regulates many important metabolic pathways in the liver, fat tissue, and hypothalamus [30,31][30][31]. It is well established that FoxO1 regulates hepatic gluconeogenesis and glycogenolysis in response to an insulin signal in the blood (Figure 2). High concentration of insulin decreases blood glucose level by promoting glucose absorption after feeding and inhibits glucose production by hepatic gluconeogenesis and glycogenolysis in the fasting state.
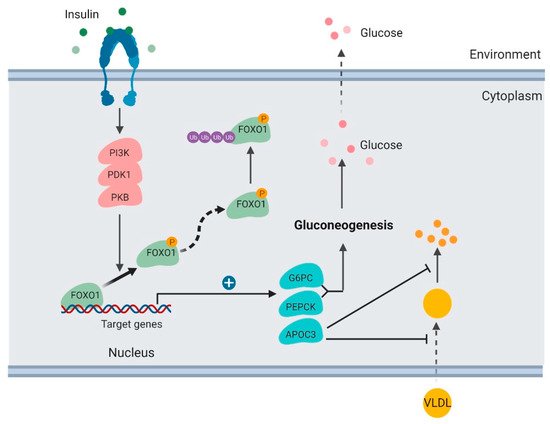
Figure 2. Regulation of glucose production and lipoprotein uptake by FOXO1 in the liver (created with BioRender.com).
In the liver, insulin activates the PI3K/PKB signaling pathway and results in FOXO1 protein phosphorylation and degradation [32]. FOXO1 TF binds and promotes transcription of glucose 6-phosphatase (G6PC) and phosphoenolpyruvate carboxykinase (PEPCK), which are key enzymes stimulating gluconeogenesis and glycogenolysis [33,34][33][34]. When FOXO1 is suppressed, transcription of G6PC and PEPCK subsequently decreases, which consequently inhibits the rates of glucose production in the liver.
It has been well established that FOXO1 is a key mediator in the signaling pathway of insulin regulating hepatic gluconeogenesis. Hepatic FOXO1 loss-of-function mutant suppresses G6PC and PEPCK expression, decreases hepatic gluconeogenesis, and improves fasting glycemia in diabetic db/db mice [32].
3.2. Lipoprotein Uptake in the Liver
Apolipoprotein C-III (ApoC3) is another downstream target of FoxO1 that functions directly in plasma triglyceride metabolism [31,35][31][35]. ApoC3 is secreted by the liver and enriched in very low-density lipoprotein (VLDL). It was reported to suppress hepatic uptake of VLDL and inhibit lipoprotein lipase [36]. ApoC3 overexpression in humans functions in atherosclerosis [37].
FOXO1 binds to the ApoC3 promoter and enhances its transcription [35] (Figure 2). FOXO1 overexpression increases hepatic ApoC3 expression and elevates plasma triglyceride levels. FOXO1 loss-of-function mutation interferes with ApoC3 expression in response to insulin stimulation. Insulin deficiency or resistance results in unrestrained ApoC3 expression and impaired triglyceride metabolism in the pathogenesis of atherosclerosis and hypertriglyceridemia.
3.3. Lipogenesis in the Liver
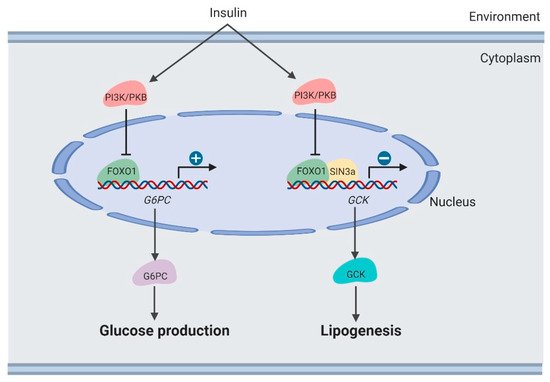
Clinically, it was observed that therapeutically decreasing blood glucose usually caused increased lipogenesis in the liver [11,38][11][38]. Lipogenesis is induced by suppressing the FoxO1-dependent inhibition of glucokinase (Gck) [39] (Figure 3). Hepatic Gck expression is associated with lipogenesis and fatty liver in humans. Increased Gck activity induced fatty liver and its metabolic consequences in humans.

Figure 3. Regulation of lipogenesis by FOXO1 in the liver (created with BioRender.com).
Moreover, SIN3 Transcription Regulator Family Member A (SIN3A) was identified as an insulin-sensitive FOXO1 corepressor of Gck. SIN3A knockout interferes with Gck expression regulated by FOXO1 in response to environmental nutrients, while not affecting expression of other genes targeted by FOXO1, such as G6PC. These results may provide the opportunity to develop selective modulators of the FoxO1 pathway, which removes adverse effects of hepatic lipogenesis in therapeutically treating diabetes [39].
3.4. Lipid Metabolism in Adipocytes
FoxO1 binds to the promoter sites of peroxisome proliferator-activated receptor gamma (PPARγ) DNA and represses its transcription [40]. In response to insulin stimulation, FOXO1 protein is degraded and subsequently unable to prevent transcription of PPARγ. Because high levels of PPARγ protein initiate adipogenesis, FoxO1 suppresses adipogenesis [41].
Moreover, the FOXO1 protein is a repressor of uncoupling protein 1 (Ucp1) gene transcription [31,42][31][42]. UCP1 protein is a mitochondrial inner membrane proton channel linked to energy use that serves as a well-known biomarker of the thermogenesis state of adipose tissues [43,44,45][43][44][45]. FoxO1 inhibition increases UCP1 expression, thereby augmenting thermogenesis and fat loss. Selectively inhibiting FTO by inhibitors decreases FOXO1 expression and reduces body weight and fat mass in a high-fat diet-induced obesity (DIO) mouse model. Suppressing FOXO1 expression increased the energy expenditure of mice. Thermogenesis in adipose tissue was induced by reduced FOXO1 expression, which was a cause of the decreased body weight and increased energy expenditure [28][29].
3.5. Feeding Behavior in the Hypothalamus
FoxO1 in the central nervous system, mainly the hypothalamus, functions directly in integrating signals from peripheral tissues and mediating feeding behavior. Insulin and leptin are well-studied nutrient signals, integrating peripheral energy status to the hypothalamus. In the arcuate nucleus (ARC) of the hypothalamus, two neuronal populations express specific feeding-related neuropeptides, including pro-opiomelanocortin (POMC) and agouti-related peptide (AgRP) [46]. POMC suppresses appetite and decreases body weight. AgRP enhances food intake and increases body weight [47,48][47][48].
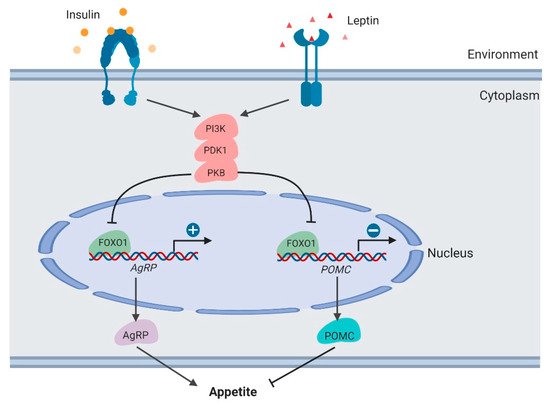
The PI3K/PDK1/PKB–FoxO1 signaling axis functions to integrate leptin and insulin signals to regulate POMC and AgRP secretion [49,50,51][49][50][51] (Figure 4). As the downstream target of these kinases, the FOXO1 protein is phosphorylated and inactivated in neurons, thus promoting POMC transcription and suppressing AgRP transcription. In mouse models, PDK1 knockout in POMC neurons suppresses FOXO1 phosphorylation and degradation, thus suppressing POMC transcription. By contrast, in AgRP neurons, PDK1 deletion results in enhanced AgRP expression and increased food intake. In the PDK1-knockout mouse models, FoxO1 inactivation rescues these phenotypes, which demonstrates the crucial roles of FoxO1 in regulating feeding behavior.

Figure 4. Regulation of appetite by FOXO1 in the hypothalamus (created with BioRender.com).
Furthermore, in a DIO rat model, the mechanism of FoxO1 in regulating feeding behavior was confirmed [52]. In these rats, because of insulin resistance, the PI3K/PDK1/PKB-FoxO1 regulatory axis was damaged, resulting in increasing the amount of FOXO1 protein and food intake. However, the intracerebroventricular (i.c.v.) micro-infusion of FoxO1-antisense oligonucleotides (FoxO1-ASOs) largely decreased the hypothalamic FoxO1 expression, food intake, and body weight gain. These suggested that pharmacological inactivation of FoxO1 may be used to suppress appetite and treat obesity.
References
- Anderson, M.J.; Viars, C.S.; Czekay, S.; Cavenee, W.K.; Arden, K.C. Cloning and characterization of three human forkhead genes that comprise an fkhr-like gene subfamily. Genomics 1998, 47, 187–199.
- Carter, M.E.; Brunet, A. Foxo transcription factors. Curr. Biol. 2007, 17, R113–R114.
- Galili, N.; Davis, R.J.; Fredericks, W.J.; Mukhopadhyay, S.; Rauscher, F.J., III; Emanuel, B.S.; Rovera, G.; Barr, F.G. Fusion of a fork head domain gene to pax3 in the solid tumour alveolar rhabdomyosarcoma. Nat. Genet. 1993, 5, 230–235.
- Boura, E.; Silhan, J.; Herman, P.; Vecer, J.; Sulc, M.; Teisinger, J.; Obsilova, V.; Obsil, T. Both the n-terminal loop and wing w2 of the forkhead domain of transcription factor foxo4 are important for DNA binding. J. Biol. Chem. 2007, 282, 8265–8275.
- Brent, M.M.; Anand, R.; Marmorstein, R. Structural basis for DNA recognition by foxo1 and its regulation by posttranslational modification. Structure 2008, 16, 1407–1416.
- Psenakova, K.; Kohoutova, K.; Obsilova, V.; Ausserlechner, M.J.; Veverka, V.; Obsil, T. Forkhead domains of foxo transcription factors differ in both overall conformation and dynamics. Cells 2019, 8, 966.
- Avila-Flores, A.; Arranz-Nicolas, J.; Merida, I. Transcriptional activity of foxo transcription factors measured by luciferase assays. Methods Mol. Biol. 2019, 1890, 91–102.
- Linardic, C.M. Pax3-foxo1 fusion gene in rhabdomyosarcoma. Cancer Lett. 2008, 270, 10–18.
- Missiaglia, E.; Williamson, D.; Chisholm, J.; Wirapati, P.; Pierron, G.; Petel, F.; Concordet, J.P.; Thway, K.; Oberlin, O.; Pritchard-Jones, K.; et al. Pax3/foxo1 fusion gene status is the key prognostic molecular marker in rhabdomyosarcoma and significantly improves current risk stratification. J. Clin. Oncol. 2012, 30, 1670–1677.
- Xing, Y.Q.; Li, A.; Yang, Y.; Li, X.X.; Zhang, L.N.; Guo, H.C. The regulation of foxo1 and its role in disease progression. Life Sci. 2018, 193, 124–131.
- Dong, X.C. Foxo transcription factors in non-alcoholic fatty liver disease. Liver Res. 2017, 1, 168–173.
- Eijkelenboom, A.; Burgering, B.M. Foxos: Signalling integrators for homeostasis maintenance. Nat. Rev. Mol. Cell Biol. 2013, 14, 83–97.
- Link, W.; Fernandez-Marcos, P.J. Foxo transcription factors at the interface of metabolism and cancer. Int. J. Cancer 2017, 141, 2379–2391.
- Maiese, K. Foxo transcription factors and regenerative pathways in diabetes mellitus. Curr. Neurovasc. Res. 2015, 12, 404–413.
- Wagatsuma, A.; Shiozuka, M.; Takayama, Y.; Hoshino, T.; Mabuchi, K.; Matsuda, R. Effects of ageing on expression of the muscle-specific e3 ubiquitin ligases and akt-dependent regulation of foxo transcription factors in skeletal muscle. Mol. Cell. Biochem. 2016, 412, 59–72.
- Pettersen, E.F.; Goddard, T.D.; Huang, C.C.; Couch, G.S.; Greenblatt, D.M.; Meng, E.C.; Ferrin, T.E. Ucsf chimera—A visualization system for exploratory research and analysis. J. Comput. Chem. 2004, 25, 1605–1612.
- Yamagata, K.; Daitoku, H.; Takahashi, Y.; Namiki, K.; Hisatake, K.; Kako, K.; Mukai, H.; Kasuya, Y.; Fukamizu, A. Arginine methylation of foxo transcription factors inhibits their phosphorylation by akt. Mol. Cell 2008, 32, 221–231.
- Biggs, W.H., III; Meisenhelder, J.; Hunter, T.; Cavenee, W.K.; Arden, K.C. Protein kinase b/akt-mediated phosphorylation promotes nuclear exclusion of the winged helix transcription factor fkhr1. Proc. Natl. Acad. Sci. USA 1999, 96, 7421–7426.
- Grabiec, A.M.; Angiolilli, C.; Hartkamp, L.M.; van Baarsen, L.G.; Tak, P.P.; Reedquist, K.A. Jnk-dependent downregulation of foxo1 is required to promote the survival of fibroblast-like synoviocytes in rheumatoid arthritis. Ann. Rheumatic Dis. 2015, 74, 1763–1771.
- Yuan, Z.; Lehtinen, M.K.; Merlo, P.; Villen, J.; Gygi, S.; Bonni, A. Regulation of neuronal cell death by mst1-foxo1 signaling. J. Biol. Chem. 2009, 284, 11285–11292.
- Brunet, A.; Sweeney, L.B.; Sturgill, J.F.; Chua, K.F.; Greer, P.L.; Lin, Y.; Tran, H.; Ross, S.E.; Mostoslavsky, R.; Cohen, H.Y.; et al. Stress-dependent regulation of foxo transcription factors by the sirt1 deacetylase. Science 2004, 303, 2011–2015.
- Pramanik, K.C.; Fofaria, N.M.; Gupta, P.; Srivastava, S.K. Cbp-mediated foxo-1 acetylation inhibits pancreatic tumor growth by targeting sirt. Mol. Cancer Ther. 2014, 13, 687–698.
- Van der Heide, L.P.; Smidt, M.P. Regulation of foxo activity by cbp/p300-mediated acetylation. Trends Biochem. Sci. 2005, 30, 81–86.
- Fu, W.; Ma, Q.; Chen, L.; Li, P.; Zhang, M.; Ramamoorthy, S.; Nawaz, Z.; Shimojima, T.; Wang, H.; Yang, Y.; et al. Mdm2 acts downstream of p53 as an e3 ligase to promote foxo ubiquitination and degradation. J. Biol. Chem. 2009, 284, 13987–14000.
- Huang, H.; Tindall, D.J. Regulation of foxo protein stability via ubiquitination and proteasome degradation. Biochim. Biophys. Acta 2011, 1813, 1961–1964.
- Vogt, P.K.; Jiang, H.; Aoki, M. Triple layer control: Phosphorylation, acetylation and ubiquitination of foxo proteins. Cell Cycle 2005, 4, 908–913.
- Dominissini, D.; Moshitch-Moshkovitz, S.; Schwartz, S.; Salmon-Divon, M.; Ungar, L.; Osenberg, S.; Cesarkas, K.; Jacob-Hirsch, J.; Amariglio, N.; Kupiec, M.; et al. Topology of the human and mouse m6a rna methylomes revealed by m6a-seq. Nature 2012, 485, 201–206.
- Meyer, K.D.; Saletore, Y.; Zumbo, P.; Elemento, O.; Mason, C.E.; Jaffrey, S.R. Comprehensive analysis of mrna methylation reveals enrichment in 3′ utrs and near stop codons. Cell 2012, 149, 1635–1646.
- Peng, S.; Xiao, W.; Ju, D.; Sun, B.; Hou, N.; Liu, Q.; Wang, Y.; Zhao, H.; Gao, C.; Zhang, S.; et al. Identification of entacapone as a chemical inhibitor of fto mediating metabolic regulation through foxo1. Sci. Transl. Med. 2019, 11, eaau7116.
- Henriques, V.; Machado, S.; Link, W.; Ferreira, B.I. Monitoring the transcriptional activity of foxo transcription factors by analyzing their target genes. Methods Mol. Biol. 2019, 1890, 103–113.
- Nakae, J.; Cao, Y.; Oki, M.; Orba, Y.; Sawa, H.; Kiyonari, H.; Iskandar, K.; Suga, K.; Lombes, M.; Hayashi, Y. Forkhead transcription factor foxo1 in adipose tissue regulates energy storage and expenditure. Diabetes 2008, 57, 563–576.
- Altomonte, J.; Richter, A.; Harbaran, S.; Suriawinata, J.; Nakae, J.; Thung, S.N.; Meseck, M.; Accili, D.; Dong, H. Inhibition of foxo1 function is associated with improved fasting glycemia in diabetic mice. Am. J. Physiol. Endocrinol. Metabol. 2003, 285, E718–E728.
- Matsumoto, M.; Pocai, A.; Rossetti, L.; Depinho, R.A.; Accili, D. Impaired regulation of hepatic glucose production in mice lacking the forkhead transcription factor foxo1 in liver. Cell Metabol. 2007, 6, 208–216.
- Wu, Y.; Pan, Q.; Yan, H.; Zhang, K.; Guo, X.; Xu, Z.; Yang, W.; Qi, Y.; Guo, C.A.; Hornsby, C.; et al. Novel mechanism of foxo1 phosphorylation in glucagon signaling in control of glucose homeostasis. Diabetes 2018, 67, 2167–2182.
- Altomonte, J.; Cong, L.; Harbaran, S.; Richter, A.; Xu, J.; Meseck, M.; Dong, H.H. Foxo1 mediates insulin action on apoc-iii and triglyceride metabolism. J. Clin. Investig. 2004, 114, 1493–1503.
- Mendivil, C.O.; Zheng, C.; Furtado, J.; Lel, J.; Sacks, F.M. Metabolism of very-low-density lipoprotein and low-density lipoprotein containing apolipoprotein c-iii and not other small apolipoproteins. Arterioscler. Thromb. Vasc. Biol. 2010, 30, 239–245.
- Bobik, A. Apolipoprotein ciii and atherosclerosis: Beyond effects on lipid metabolism. Circulation 2008, 118, 702–704.
- Haeusler, R.A.; Hartil, K.; Vaitheesvaran, B.; Arrieta-Cruz, I.; Knight, C.M.; Cook, J.R.; Kammoun, H.L.; Febbraio, M.A.; Gutierrez-Juarez, R.; Kurland, I.J.; et al. Integrated control of hepatic lipogenesis versus glucose production requires foxo transcription factors. Nat. Commun. 2014, 5, 5190.
- Langlet, F.; Haeusler, R.A.; Linden, D.; Ericson, E.; Norris, T.; Johansson, A.; Cook, J.R.; Aizawa, K.; Wang, L.; Buettner, C.; et al. Selective inhibition of foxo1 activator/repressor balance modulates hepatic glucose handling. Cell 2017, 171, 824–835.
- Farmer, S.R. The forkhead transcription factor foxo1: A possible link between obesity and insulin resistance. Mol. Cell 2003, 11, 6–8.
- Nakae, J.; Kitamura, T.; Kitamura, Y.; Biggs, W.H., III; Arden, K.C.; Accili, D. The forkhead transcription factor foxo1 regulates adipocyte differentiation. Dev. Cell 2003, 4, 119–129.
- Kita, M.; Nakae, J.; Kawano, Y.; Asahara, H.; Takemori, H.; Okado, H.; Itoh, H. Zfp238 regulates the thermogenic program in cooperation with foxo1. iScience 2019, 12, 87–101.
- Harms, M.; Seale, P. Brown and beige fat: Development, function and therapeutic potential. Nat. Med. 2013, 19, 1252–1263.
- Inagaki, T.; Sakai, J.; Kajimura, S. Transcriptional and epigenetic control of brown and beige adipose cell fate and function. Nat. Rev. Mol. Cell Biol. 2016, 17, 480–495.
- Wu, J.; Cohen, P.; Spiegelman, B.M. Adaptive thermogenesis in adipocytes: Is beige the new brown? Genes Dev. 2013, 27, 234–250.
- Varela, L.; Horvath, T.L. Leptin and insulin pathways in pomc and agrp neurons that modulate energy balance and glucose homeostasis. EMBO Rep. 2012, 13, 1079–1086.
- Belgardt, B.F.; Okamura, T.; Bruning, J.C. Hormone and glucose signalling in pomc and agrp neurons. J. Physiol. 2009, 587, 5305–5314.
- Breen, T.L.; Conwell, I.M.; Wardlaw, S.L. Effects of fasting, leptin, and insulin on agrp and pomc peptide release in the hypothalamus. Brain Res. 2005, 1032, 141–148.
- Belgardt, B.F.; Husch, A.; Rother, E.; Ernst, M.B.; Wunderlich, F.T.; Hampel, B.; Klockener, T.; Alessi, D.; Kloppenburg, P.; Bruning, J.C. Pdk1 deficiency in pomc-expressing cells reveals foxo1-dependent and -independent pathways in control of energy homeostasis and stress response. Cell Metab. 2008, 7, 291–301.
- Cao, Y.; Nakata, M.; Okamoto, S.; Takano, E.; Yada, T.; Minokoshi, Y.; Hirata, Y.; Nakajima, K.; Iskandar, K.; Hayashi, Y.; et al. Pdk1-foxo1 in agouti-related peptide neurons regulates energy homeostasis by modulating food intake and energy expenditure. PLoS ONE 2011, 6, e18324.
- Iskandar, K.; Cao, Y.; Hayashi, Y.; Nakata, M.; Takano, E.; Yada, T.; Zhang, C.; Ogawa, W.; Oki, M.; Chua, S., Jr.; et al. Pdk-1/foxo1 pathway in pomc neurons regulates pomc expression and food intake. Am. J. Physiol. Endocrinol. Metab. 2010, 298, E787–E798.
- Ropelle, E.R.; Pauli, J.R.; Prada, P.; Cintra, D.E.; Rocha, G.Z.; Moraes, J.C.; Frederico, M.J.; da Luz, G.; Pinho, R.A.; Carvalheira, J.B.; et al. Inhibition of hypothalamic foxo1 expression reduced food intake in diet-induced obesity rats. J. Physiol. 2009, 587, 2341–2351.
More
