You're using an outdated browser. Please upgrade to a modern browser for the best experience.
Please note this is a comparison between Version 2 by Vicky Zhou and Version 1 by Sheng-Fan Wang.
Animal lectins are proteins with carbohydrate recognition activity. Galectins, the β-galactoside binding lectins, are expressed in various cells and have been reported to regulate several immunological and physiological responses. Recently, some galectins have been reported to regulate some viral infections, including influenza A virus (IAV); however, the mechanism is still not fully understood.
- galectins
- influenza A virus
- carbohydrate recognition domain
- anti-influenza
1. Introduction
Influenza is an acute, contagious infectious disease caused by the influenza virus. Influenza viruses belong to the Orthomyxoviridae family, with six to eight segments of linear negative-sense, single-stranded RNA and comprise a family of four distinct viruses (influenza A, B, C, and D viruses) [1]. Among them, influenza A and B viruses are the pathogens causing seasonal influenza disease. The seasonal influenza virus epidemics are estimated to cause 3–5 million cases of severe infection and result in 290,000–650,000 deaths annually worldwide [2][3]. Higher mortality rates are seen in the elderly over 65 years old and children under 5 years old, as well as people in developing countries [4]. The burden for seasonal influenza epidemics is substantial and is significantly increased during influenza pandemics, such as a more recent, swine-originated H1N1 influenza pandemic in 2009 [5].
Great improvement has been achieved in the treatment of influenza, especially for patients with severe influenza infection. To date, the most effective strategies for the prevention and control of influenza disease are vaccination and antiviral therapy. Although influenza vaccine and antiviral drugs greatly reduced influenza outbreaks, there are still many limitations to these two approaches [6][7][8]. The current influenza vaccine can be either trivalent or quadrivalent, which may contain the strains of seasonal circulating influenza A (H1N1, H3N2) and influenza B (Yamagata or Victoria lineage). A substantial challenge for influenza vaccine is the continuous viral antigenic changes and variations, which may lead to vaccine mismatch that results in the reduction in the vaccine’s effectiveness (protection rate from 10 to 60%) [9][10]. As to influenza antiviral drugs, the major candidates are the inhibitors that can block NA activity and M2 protein function to ameliorate influenza release and viral uncoating during the influenza virus life cycle. The most common clinical treatment of influenza are NA inhibitors including, oseltamivir, zanamivir, and peramivir, as well as the M2 proton channel blockers including amantadine and rimantadine. Unfortunately, several emerging strains of influenza A virus, such as the 2009 H1N1 influenza virus and some avian H5N1 or H7N9 isolates, were reported to be naturally carrying drug-mutation genes, which leads to resistance against these drugs [11].
Lectins are a group of proteins with carbohydrate recognition activity, and they are characterized into several families based on their different cellular locations and their specificities for a variety of carbohydrate structures according to the features of their carbohydrate recognition domain (CRD) binding preference. Interaction between lectins and their recognition glycans mainly relies on their CRD binding to certain particular oligosaccharides expressed on cellular or microbial surfaces. Lectins have been categorized into many families based on their conserved structure of sequence motifs for sugar-binding and carbohydrate specificities such as calnexin, DC-SIGN, L-SIGN, mannose-6-phosphate receptors (MPRs), siglecs, galectins, and intelectins [12][13]. Recently, several lectins have been reported to participate in the regulation of virus infection and replication [14][15].
Galectins, previously categorized as S-type lectin, have a preference to bind β-galactoside sugars, such as N-acetyllactosamine (Galβ1-3GlcNAc or Galβ1-4GlcNAc), which are presented in N-linked and O-linked glycoproteins [16][17]. Galectins are the most conserved and ubiquitous lectin family, detected from protists to mammals. The first galectin was identified and characterized from the electric organs of the electric eel, Electrophorus electricus [18], and since then, members of the galectin family have been found in mammals, birds, amphibians, fish, nematodes, sponges, and some fungi [19][20]. Galectins have been reported to regulate various cellular or immunological functions [21]. Galectins are reported to be the pattern recognition receptors (PRRs) to recognize microbial invasion and regulate innate immune responses [22]. The basic structure of a galectin contains a carbohydrate recognition domain (CRD) (about 130 amino acids) that connects to a tandem repeat domain. Currently, galectins have been categorized into three main types—prototype, chimera type, and tandem-repeat type. The prototype galectins (Gal-1, Gal-2, Gal-5, Gal-7, Gal-10 Gal-11, Gal-13, Gal-14, and Gal-15) contain one CRD and are either monomers or noncovalent homodimers; the unique chimera-type galectin (Gal-3) contains a rare tandem that repeats of proline and glycine-rich short stretches fused onto the CRD; and tandem-repeat galectins (Gal-4, Gal-6, Gal-8, Gal-9, and Gal-12) contain two distinct CRDs, in tandem, connected by a linker peptide. Currently, 15 galectins have been identified in mammals, whereas 12 galectins are detected in humans [21][23] (Figure 1).
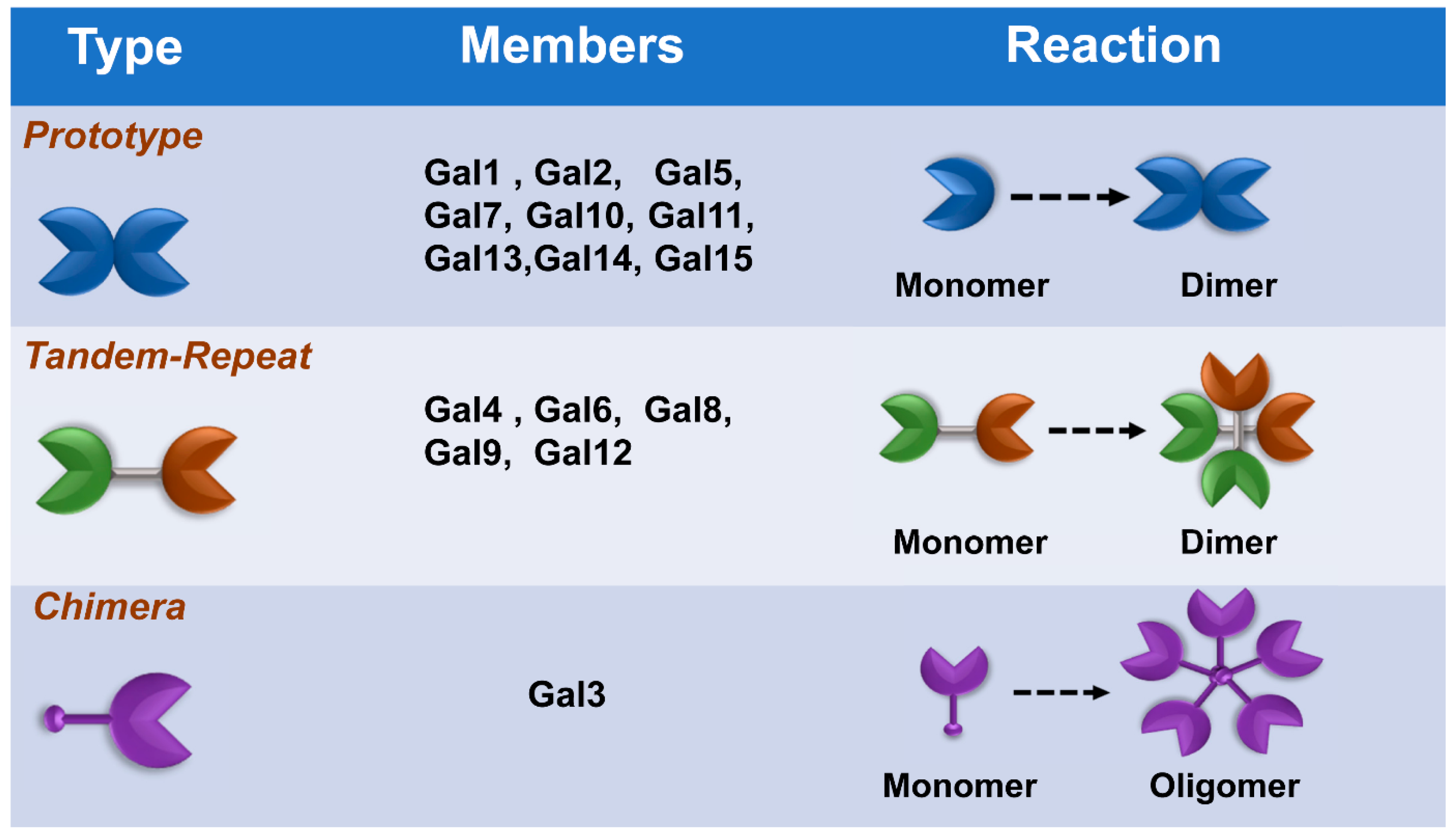 Figure 1. Classification of galectins. According to the number and arrangement of the carbohydrate recognition domains (CRDs), galectin family members are classified into three main types: prototype, chimera type, and tandem-repeat type. Some galectins can self-associate into dimers or oligomers.
Figure 1. Classification of galectins. According to the number and arrangement of the carbohydrate recognition domains (CRDs), galectin family members are classified into three main types: prototype, chimera type, and tandem-repeat type. Some galectins can self-associate into dimers or oligomers.
Recently, galectins have been reported to participate in the regulation of several viral infections and replications, such as HIV-1, enterovirus, HSV-1, adenovirus, and dengue viruses. Some reports indicated that galectins have abilities to ameliorate influenza virus infection; however, the details of the role and regulatory mechanism are still not fully understood [17][24][25]. Accordingly, this study performs a systemic literature review to analyze and summarize the potential anti-influenza function of galectins.
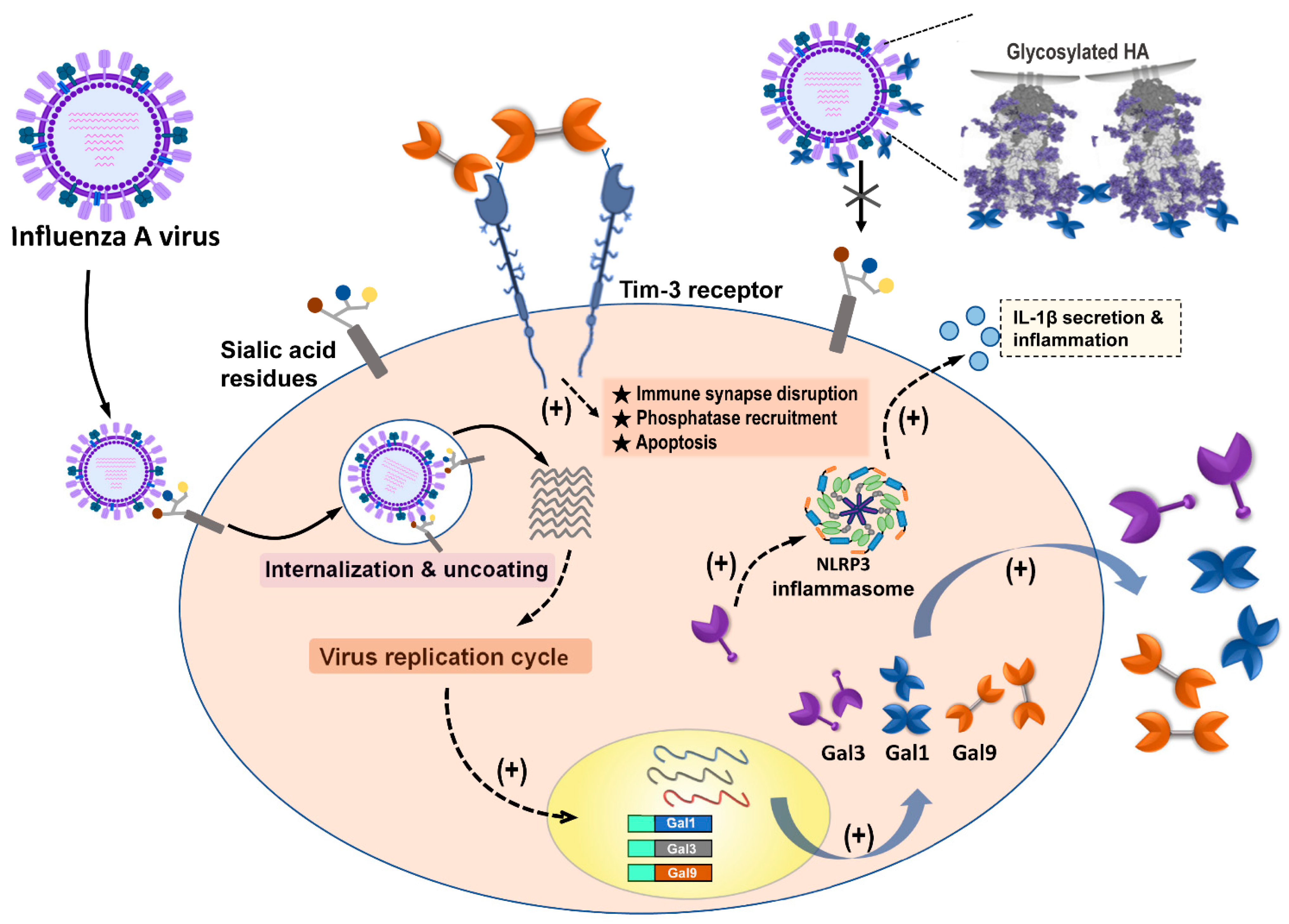 Figure 42. Summary of galectins against influenza A virus infection. Influenza A virus (IAV) infection triggered the induction and secretion of galectin-1 (Gal-1), galectin-3 (Gal-3), and galectin-9 (Gal-9). Secreted Gal-1 could bind to HA glycoprotein of IAVs and further blocking IAV interaction with sialic acid receptors expressed on the cells. IAV infection upregulated endogenous Gal-3, which could induce NLRP3 inflammasome activation, as well as IL-1β secretion, to result in inflammation occurrence. The secreted Gal-9 could interact with Tim-3 expressing cells. Gal-9/Tim-3 interaction triggered Tim-3 downstream signaling and induced apoptosis.
Figure 42. Summary of galectins against influenza A virus infection. Influenza A virus (IAV) infection triggered the induction and secretion of galectin-1 (Gal-1), galectin-3 (Gal-3), and galectin-9 (Gal-9). Secreted Gal-1 could bind to HA glycoprotein of IAVs and further blocking IAV interaction with sialic acid receptors expressed on the cells. IAV infection upregulated endogenous Gal-3, which could induce NLRP3 inflammasome activation, as well as IL-1β secretion, to result in inflammation occurrence. The secreted Gal-9 could interact with Tim-3 expressing cells. Gal-9/Tim-3 interaction triggered Tim-3 downstream signaling and induced apoptosis.
2. The Antiviral Role of Galectins toward Influenza A Virus Infection
Our results indicate that among galectin members, only Gal-1, Gal-3, and Gal-9 were reported to have abilities to regulate influenza infection and replication via directly binding to glycosylated influenza HA or indirectly enhancing the immune response against IAV invasion (Figure 4Figure 2) (Table 1). We also noted that out of the other type of influenza virus, only IAV had studies that reported its interaction, correlation, and modulation with galectins. Most studies provided lines of evidence denoting that expression or treatment with Gal-1 and Gal-9 ameliorated IAV infection via blocking viral binding and enhancing T-cell immune responses, respectively. The Gal-3 expression also participated in the amelioration of IAV infection; however, there were a few studies that offered data showing that Gal-3 had promotion capabilities during IAV and S. pneumoniae coinfection in vitro and in vivo models (Table 1). Figure 42. Summary of galectins against influenza A virus infection. Influenza A virus (IAV) infection triggered the induction and secretion of galectin-1 (Gal-1), galectin-3 (Gal-3), and galectin-9 (Gal-9). Secreted Gal-1 could bind to HA glycoprotein of IAVs and further blocking IAV interaction with sialic acid receptors expressed on the cells. IAV infection upregulated endogenous Gal-3, which could induce NLRP3 inflammasome activation, as well as IL-1β secretion, to result in inflammation occurrence. The secreted Gal-9 could interact with Tim-3 expressing cells. Gal-9/Tim-3 interaction triggered Tim-3 downstream signaling and induced apoptosis.
Figure 42. Summary of galectins against influenza A virus infection. Influenza A virus (IAV) infection triggered the induction and secretion of galectin-1 (Gal-1), galectin-3 (Gal-3), and galectin-9 (Gal-9). Secreted Gal-1 could bind to HA glycoprotein of IAVs and further blocking IAV interaction with sialic acid receptors expressed on the cells. IAV infection upregulated endogenous Gal-3, which could induce NLRP3 inflammasome activation, as well as IL-1β secretion, to result in inflammation occurrence. The secreted Gal-9 could interact with Tim-3 expressing cells. Gal-9/Tim-3 interaction triggered Tim-3 downstream signaling and induced apoptosis.Table 1. Summary of the antiviral roles of galectins against influenza virus infection.
| Galectin | Antiviral Effects | Target Virus | Ref. | |
|---|---|---|---|---|
| Gal-1 directly binds to the envelope glycoproteins of influenza virus and constrain the viral hemagglutination activity and infectivity. | A/WSN/1933(H1N1) | [26] | ||
| Galectin-1 | Recombinant Gal-1 (rGal-1) treatment reduced mice fatality via mediating the expression of cytokines and chemokines. | 2009 influenza A H1N1 subtype (H1N1pdm09) | [25] | |
| Gal-1 participated in regulation of cytopathic processes by H1N1pdm09 virus to induce an arrest of the cell cycle at the G0/G1 phase. | H1N1pdm09 | [27] | ||
| Gal-1 expression was correlated with the differential susceptibility to H7N9 influenza via extracellular matrix (ECM)-receptor interaction and mitogen-activated protein kinase (MAPK) signaling. | Human H7N9 isolates | [28] | ||
| IAVs and | S. pneumoniae | coinfection induced the secretion of Gal-1 to the epithelial cell surface and further modulated the expression of SOCS1 and RIG1 and activate ERK, AKT, or JAK/STAT1 signaling pathways. | H1N1pdm09 | [29] |
| Galectin-3 | Aloe-emodin treatment ameliorated influenza H1N1 virus infection via up-regulation of Gal-3 expression to further trigger antiviral genes expression | A/Taiwan/CMUH01/2007(H1N1) | [30] | |
| Gal-3 enhances effects of H5N1 promoting host inflammatory response by up-regulating IL-1β via NLRP3. | A/Vietnam/1204/03 | [31] | ||
| IAVs and | S. pneumoniae | coinfection induced the secretion of Gal-3 to the epithelial cell surface and further modulated the expression of SOCS1 and RIG1, and activate ERK, AKT, or JAK/STAT1 signaling pathways. | H1N1pdm09 | [29] |
| Gal-3 preferred binding to desialylated multivalent glycoligands. | A/PuertoRico/08/1934 (H1N1) | [32] | ||
| Galectin-9 | Gal-9 inhibited the infection of IAVs via Gal-9 binding to influenza virus particles to inhibit virus attachment. | A/Puerto Rico/8/34 (H1N1); Aichi/2/68 (H3N2); A/Hong Kong/483/97 (H5N1) | [33] | |
| Virus-specific CD8 T cells upregulate Tim-3 expression and Gal-9/Tim-3 interaction induce cell apoptosis after IAV infection from in vitro and ex vivo assays. | HK/×31 (H3N2) A/Puerto Rico/8/34 (H1N1)) | [34] | ||
| Influenza virus infection induces plasma Gal-9 expression, suggesting Gal-9 as a possible biomarker for influenza. | Seasonal influenza virus | [35] |
Galectins were initially discovered in 197, based on their galactoside-binding activity. Galectins reside in the cytosol or nucleus for the majority of their lifetime, and they reach their galactoside ligands only after nonclassical secretion that bypasses the Golgi apparatus [18]. Accordingly, many previous studies focused on studying the regulatory functions of CRD of galectins and their interaction with glycan or glycoconjugates expressed on the cell surface. However, instead of glycan–CRD interaction, the endogenous functions of galectins which exert their regulatory function via protein–protein interaction are gradually gaining attention. For example, Chen et al. reported that endogenous Gal-3 expression induced pulmonary inflammasome during H5N1 avian influenza infection via binding to NLRP3 and ASC and further enhancing ASC oligomerization and NLRP3 inflammasome activation [31]. This regulatory effect, induced by endogenous Gal-3 on the H5N1 virus, suggests a protein–protein interaction.
Galectins are known to be involved in various biological and biophysical regulations, including defense against microorganisms [36]. Recently, galectins have been recognized as modulators and pattern recognition receptors (PRRs) in response to virus or bacterial attack [24][36][37]. Our study indicates that only a few galectins were reported to participate in the regulation of IAV infection, replication, and propagation. Despite there being 12 animal galectins identified in humans, only Gal-1, Gal-3, and Gal-9, characterized as the prototype, chimera, and tandem-repeat type of galectins, respectively, have been reported to have regulatory effects on IAVs. Each type of galectin family contains several members and each member in the same family are proposed to have similarities in sequence and structure as well as comparable regulatory capabilities [38]. Nevertheless, they might be some differences owing to the 20–40% identity of amino acids in the CRDs among the different galectins and 80–90% identity of the same galectin from different mammalian species [39]. We, therefore, suggest that there might be more galectins with regulatory capabilities toward IAV infection; hence, further investigation is required. Furthermore, while galectins have been wieldy studied in various fields, the discovery of galectins in virus research is still in its initial phase.
To date, the conventional strategy for influenza treatment is the usage of antiviral drugs [11][40]. The two major antiviral drugs that are used for the clinical treatment of influenza include NA and M2 protein inhibitors, which inhibit NA activities and block M2 ion channels, consequently inhibiting virus budding and the vRNP release [11]. Unfortunately, the occurrence of influenza drug resistance causes a major problem for this influenza treatment strategy. Reports indicated that approximately 45% of all influenza A subtypes in circulation, globally, were resistant to the adamantanes by 2013 (>69% of H1 subtypes and 43% of H3 subtypes) [11]. Resistance to NA inhibitor shows a lesser increase in comparison with the adamantanes. Prior to 2007, oseltamivir resistance was 1–5% detected in clinical practice. During the 2008–2009 influenza season, several countries reported isolating high oseltamivir-resistant strains [41]. Fortunately, the 2009 pandemic (pdm09) influenza A H1N1 strain emerged globally, and most infected cases showed susceptibility to oseltamivir treatment [42]. Based on this information, we recommend that there is a demand for the development of a novel anti-influenza strategy to overcome influenza-drug resistance; thus, we propose that galectins may have the potential to be applied as an alternative anti-influenza treatment strategy.
Furthermore, our results indicated that Gal-1, Gal-3, and Gal-9 were upregulated after the IAV invasion. These galectins are ubiquitously expressed in various cells and are detected intracellularly, in the cytoplasm and the nucleus, as well as extracellularly, outside the environment. In addition, the influenza virion contains abundant glycosylated HA and NA on its envelope (about 350 to 400 HA trimers and 50 neuraminidase tetramers) [43]. The glycosylation of HA and NA were shown in various degrees among different subtypes or strains of the influenza virus. The HA molecules have glycosylation sites ranging between 5 and 11, whereas the NA molecule has three to five potential glycosylation sites [44][45]. Gal-1, Gal-3, and Gal-9 were all reported to bind to IAVs via the interaction of its CRD with viral glycans, but only Gal-1 and Gal-9 were reported to result in the inhibition of HA binding to sialic acid receptors on host cells. However, Gal-3 did not show the enhancing effect on influenza attachment and internalization via extracellularly adding route [31]. Nevertheless, the crosslink and multivalent activity also displayed by Gal-3 when interacting with glycan might suggest that comparable CRD–glycan interaction still has a chance to result in a different regulatory outcome. However, the detailed mechanism of this phenomenon remains unclear.
Here, we reported that Gal-3 regulated avian H5N1 virus infection via promotion of NLRP3 inflammasome activation to enhance pulmonary inflammation [31]. The avian H5N1 virus has been reported to be more virulent than the seasonal influenza virus and could induce severe pneumonia, immune dysregulation, and cytokine storm [46]. However, the detailed mechanism of avian H5N1-induced pathogenicity and higher mortality remains not fully understood. During influenza virus infection, NLR family members are known to play important roles to regulate antiviral responses, especially for NLRP3 inflammasome since it has been reported to mediate several virus infections via promoting antiviral immune responses [47]. Except for IAVs, several studies have demonstrated the essential role of endogenous Gal-3 in infection-induced inflammatory response against either virus or bacteria invasion via inducing neutrophil infiltration or proinflammatory cytokine production such as IL-1β, TNF-α, and IFN-γ [17]. Accordingly, we suggest that Gal-3 plays a role in the regulation of microbial infection by increasing inflammatory response.
Similarly, Gal-9 was also reported to be upregulated during IAV infection and exerted anti-influenza effects [33]. Our results denoted that Gal-9 and Tim-3 interaction resulted in the amelioration of IAV infection and replication. Previous reports indicated that Gal-9 binding to Tim-3 on T cells could limit the extent of immunopathological lesions in autoimmunity, as well as in some chronic infections [48]. From the immunological viewpoint, the host immune response to virus infection needs precise regulation to minimize tissue damage while still achieving defense. However, some bystander tissue damage usually occurs due to several host defenses, enhancing inflammatory reactions and destroying nearby cells. Gal-9 is reported to play an immunomodulatory role in various microbial infections via interaction with its receptor Tim3, suggesting its regulatory effect on virus mainly through inducing immunopathogenesis [49]. Although a report indicated that Gal-9 inhibitory effect on IAVs occurs by binding to the virus particle [33], currently, most studies supported that Gal-9/Tim-3 interaction and downstream signaling is the key factor to modulate several virus infections, including IAVs [49][34]. However, Gal-9/Tim-3 signaling was not only found in the macrophage since this interaction was also detected in NK, CD4+, CD8+, and FoxP3+ regulatory T cells. We, therefore, suggest that further investigations are required to understand Gal-9/Tim-3 interaction signaling and how Gal-9 affects IAV infection through regulating different immune cells.
Discovering novel or even alternative therapeutic strategies is necessary to overcome the limitations of the current use of conventional antiviral drugs and vaccines against influenza viruses. In this study, we suggest that galectins have anti-influenza capabilities and could be a potential candidate to develop an alternative influenza treatment and prophylactic control use.
References
- Bouvier, N.M.; Palese, P. The biology of influenza viruses. Vaccine 2008, 26, D49–D53.
- Erbelding, E.J.; Post, D.J.; Stemmy, E.J.; Roberts, P.C.; Augustine, A.D.; Ferguson, S.; Paules, C.I.; Graham, B.S.; Fauci, A.S. A universal influenza vaccine: The strategic plan for the National Institute of Allergy and Infectious Diseases. J. Infect. Dis. 2018, 218, 347–354.
- WHO. Influenza (Seasonal). Available online: (accessed on 6 November 2018).
- Iuliano, A.D.; Roguski, K.M.; Chang, H.H.; Muscatello, D.J.; Palekar, R.; Tempia, S.; Cohen, C.; Gran, J.M.; Schanzer, D.; Cowling, B.J.; et al. Estimates of global seasonal influenza-associated respiratory mortality: A modelling study. Lancet 2018, 391, 1285–1300.
- Van Kerkhove, M.D.; Hirve, S.; Koukounari, A.; Mounts, A.W.; H1N1pdm Serology Working Group. Estimating age-specific cumulative incidence for the 2009 influenza pandemic: A meta-analysis of A(H1N1)pdm09 serological studies from 19 countries. Influenza Other Respir. Viruses 2013, 7, 872–886.
- Goodwin, K.; Viboud, C.; Simonsen, L. Antibody response to influenza vaccination in the elderly: A quantitative review. Vaccine 2006, 24, 1159–1169.
- Soema, P.C.; Kompier, R.; Amorij, J.P.; Kersten, G.F. Current and next generation influenza vaccines: Formulation and production strategies. Eur. J. Pharm. Biopharm. 2015, 94, 251–263.
- Lewnard, J.A.; Cobey, S. Immune history and influenza vaccine effectiveness. Vaccines 2018, 6, 28.
- CDC. Seasonal Flu Vaccine Effectiveness Studies. Available online: (accessed on 11 December 2020).
- Grohskopf, L.A.; Alyanak, E.; Broder, K.R.; Blanton, L.H.; Fry, A.M.; Jernigan, D.B.; Atmar, R.L. Prevention and control of seasonal influenza with vaccines: Recommendations of the advisory committee on immunization practices—United States, 2020–21 influenza season. MMWR Recomm. Rep. 2020, 69, 1–24.
- Hussain, M.; Galvin, H.D.; Haw, T.Y.; Nutsford, A.N.; Husain, M. Drug resistance in influenza A virus: The epidemiology and management. Infect. Drug Resist. 2017, 10, 121–134.
- Gupta, G.S.G.A.; Gupta, R.K. Lectins: An overview. In Animal Lectins: Forms, Functions and Clinical Applications; Gupta, G.S., Ed.; Springer: New York, NY, USA, 2012; Volume 1, p. 1108.
- Liu, Y.; Liu, J.; Pang, X.; Liu, T.; Ning, Z.; Cheng, G. The roles of direct recognition by animal lectins in antiviral immunity and viral pathogenesis. Molecules 2015, 20, 2272–2295.
- Mason, C.P.; Tarr, A.W. Human lectins and their roles in viral infections. Molecules 2015, 20, 2229–2271.
- Wang, S.F.; Huang, J.C.; Lee, Y.M.; Liu, S.J.; Chan, Y.J.; Chau, Y.P.; Chong, P.; Chen, Y.M. DC-SIGN mediates avian H5N1 influenza virus infection in cis and in trans. Biochem. Biophys. Res. Commun. 2008, 373, 561–566.
- Machala, E.A.; McSharry, B.P.; Rouse, B.T.; Abendroth, A.; Slobedman, B. Gal power: The diverse roles of galectins in regulating viral infections. J. Gen. Virol. 2019, 100, 333–349.
- Wang, W.H.; Lin, C.Y.; Chang, M.R.; Urbina, A.N.; Assavalapsakul, W.; Thitithanyanont, A.; Chen, Y.H.; Liu, F.T.; Wang, S.F. The role of galectins in virus infection—A systemic literature review. J. Microbiol. Immunol. Infect. 2020, 53, 925–935.
- Teichberg, V.I.; Silman, I.; Beitsch, D.D.; Resheff, G. A beta-D-galactoside binding protein from electric organ tissue of Electrophorus electricus. Proc. Natl. Acad. Sci. USA 1975, 72, 1383–1387.
- Houzelstein, D.; Goncalves, I.R.; Fadden, A.J.; Sidhu, S.S.; Cooper, D.N.; Drickamer, K.; Leffler, H.; Poirier, F. Phylogenetic analysis of the vertebrate galectin family. Mol. Biol. Evol. 2004, 21, 1177–1187.
- Vasta, G.R.; Ahmed, H.; Nita-Lazar, M.; Banerjee, A.; Pasek, M.; Shridhar, S.; Guha, P.; Fernandez-Robledo, J.A. Galectins as self/non-self recognition receptors in innate and adaptive immunity: An unresolved paradox. Front. Immunol. 2012, 3, 199.
- Cummings, R.D.; Liu, F.T.; Vasta, G.R. Galectins. In Essentials of Glycobiology, 3rd ed.; Varki, A., Cummings, R.D., Esko, J.D., Stanley, P., Hart, G.W., Aebi, M., Darvill, A.G., Kinoshita, T., Packer, N.H., Prestegard, J.H., et al., Eds.; Cold Spring Harbor: New York, NY, USA, 2017; pp. 469–480.
- Vasta, G.R. Roles of galectins in infection. Nat. Rev. Microbiol. 2009, 7, 424–438.
- Ayona, D.; Fournier, P.E.; Henrissat, B.; Desnues, B. Utilization of Galectins by Pathogens for Infection. Front Immunol 2020, 11, 1877.
- Yang, Z.-S.; Lin, C.-Y.; Huang, S.-W.; Wang, W.-H.; Urbina, A.N.; Tseng, S.-P.; Lu, P.-L.; Chen, Y.-H.; Wang, S.-F. Regulatory roles of galectins on influenza A virus and their potential as a therapeutic strategy. Biomed. Pharmacother. 2021, 139, 111713.
- Bao, J.; Wang, X.; Liu, S.; Zou, Q.; Zheng, S.; Yu, F.; Chen, Y. Galectin-1 ameliorates influenza A H1N1pdm09 virus-induced acute lung injury. Front. Microbiol. 2020, 11, 1293.
- Yang, M.L.; Chen, Y.H.; Wang, S.W.; Huang, Y.J.; Leu, C.H.; Yeh, N.C.; Chu, C.Y.; Lin, C.C.; Shieh, G.S.; Chen, Y.L.; et al. Galectin-1 binds to influenza virus and ameliorates influenza virus pathogenesis. J. Virol. 2011, 85, 10010–10020.
- Fang, S.; Zhang, K.; Wang, T.; Wang, X.; Lu, X.; Peng, B.; Wu, W.; Zhang, R.; Chen, S.; Zhang, R.; et al. Primary study on the lesions and specific proteins in BEAS-2B cells induced with the 2009 A (H1N1) influenza virus. Appl. Microbiol. Biotechnol. 2014, 98, 9691–9701.
- Chen, Y.; Zhou, J.; Cheng, Z.; Yang, S.; Chu, H.; Fan, Y.; Li, C.; Wong, B.H.; Zheng, S.; Zhu, Y.; et al. Functional variants regulating LGALS1 (Galectin 1) expression affect human susceptibility to influenza A(H7N9). Sci. Rep. 2015, 5, 8517.
- Nita-Lazar, M.; Banerjee, A.; Feng, C.; Vasta, G.R. Galectins regulate the inflammatory response in airway epithelial cells exposed to microbial neuraminidase by modulating the expression of SOCS1 and RIG1. Mol. Immunol. 2015, 68, 194–202.
- Li, S.-W.; Yang, T.-C.; Lai, C.-C.; Huang, S.-H.; Liao, J.-M.; Wan, L.; Lin, Y.-J.; Lin, C.-W. Antiviral activity of aloe-emodin against influenza A virus via galectin-3 up-regulation. Eur. J. Pharmacol. 2014, 738, 125–132.
- Chen, Y.J.; Wang, S.F.; Weng, I.C.; Hong, M.H.; Lo, T.H.; Jan, J.T.; Hsu, L.C.; Chen, H.Y.; Liu, F.T. Galectin-3 enhances avian H5N1 influenza A virus-induced pulmonary inflammation by promoting NLRP3 inflammasome activation. Am. J. Pathol. 2018, 188, 1031–1042.
- Wang, H.; Huang, W.; Orwenyo, J.; Banerjee, A.; Vasta, G.R.; Wang, L.X. Design and synthesis of glycoprotein-based multivalent glyco-ligands for influenza hemagglutinin and human galectin-3. Bioorg. Med. Chem. 2013, 21, 2037–2044.
- Hattori, T.A.T.; Fujioka, Y.; Maruyama, J.; Nakayama, Y.; Ohba, Y.; Niki, T.; Miyazaki, T.; Hirashima, M.; Kida, H. Inhibition of influenza A virus infection by Galectin-9. Jpn. J. Vet. Res. 2013, 61, 5–18.
- Sharma, S.; Sundararajan, A.; Suryawanshi, A.; Kumar, N.; Veiga-Parga, T.; Kuchroo, V.K.; Thomas, P.G.; Sangster, M.Y.; Rouse, B.T. T cell immunoglobulin and mucin protein-3 (Tim-3)/galectin-9 interaction regulates influenza A virus-specific humoral and CD8 T-cell responses. Proc. Natl. Acad. Sci. USA 2011, 108, 19001–19006.
- Katoh, S.; Ikeda, M.; Shimizu, H.; Mouri, K.; Obase, Y.; Kobashi, Y.; Fukushima, K.; Hirashima, M.; Oka, M. Increased levels of plasma galectin-9 in patients with influenza virus infection. Tohoku J. Exp. Med. 2014, 232, 263–267.
- Yang, R.Y.; Rabinovich, G.A.; Liu, F.T. Galectins: Structure, function and therapeutic potential. Expert Rev. Mol. Med. 2008, 10, e17.
- Vasta, G.R. Galectins as pattern recognition receptors: Structure, function, and evolution. Adv. Exp. Med. Biol. 2012, 946, 21–36.
- Arthur, C.M.; Baruffi, M.D.; Cummings, R.D.; Stowell, S.R. Evolving mechanistic insights into galectin functions. Methods Mol. Biol. 2015, 1207, 1–35.
- Oda, Y.; Herrmann, J.; Gitt, M.A.; Turck, C.W.; Burlingame, A.L.; Barondes, S.H.; Leffler, H. Soluble lactose-binding lectin from rat intestine with two different carbohydrate-binding domains in the same peptide chain. J. Biol. Chem. 1993, 268, 5929–5939.
- Plans, P. Recommendations for the prevention and treatment of influenza using antiviral drugs based on cost-effectiveness. Expert Rev. Pharmacoecon. Outcomes Res. 2008, 8, 563–573.
- McKimm-Breschkin, J.L. Influenza neuraminidase inhibitors: Antiviral action and mechanisms of resistance. Influenza Other Respir. Viruses 2013, 7, 25–36.
- Matos, A.R.; Resende, P.C.; Miranda, M.D.; Garcia, C.C.; Caetano, B.C.; Lopes, J.C.O.; Debur, M.C.; Cury, A.L.F.; Vianna, L.A.; Lima, M.C.; et al. Susceptibility of Brazilian influenza A(H1N1)pdm09 viruses to neuraminidase inhibitors in the 2014–2016 seasons: Identification of strains bearing mutations associated with reduced inhibition profile. Antivir. Res. 2018, 154, 35–43.
- Matrosovich, M.; Klenk, H.D. Natural and synthetic sialic acid-containing inhibitors of influenza virus receptor binding. Rev. Med. Virol. 2003, 13, 85–97.
- Skehel, J.J.; Stevens, D.J.; Daniels, R.S.; Douglas, A.R.; Knossow, M.; Wilson, I.A.; Wiley, D.C. A carbohydrate side chain on hemagglutinins of Hong Kong influenza viruses inhibits recognition by a monoclonal antibody. Proc. Natl. Acad. Sci. USA 1984, 81, 1779–1783.
- Wilson, I.A.; Skehel, J.J.; Wiley, D.C. Structure of the haemagglutinin membrane glycoprotein of influenza virus at 3 A resolution. Nature 1981, 289, 366–373.
- To, K.K.; Ng, K.H.; Que, T.L.; Chan, J.M.; Tsang, K.Y.; Tsang, A.K.; Chen, H.; Yuen, K.Y. Avian influenza A H5N1 virus: A continuous threat to humans. Emerg. Microbes Infect. 2012, 1, e25.
- WHO. Influenza (Avian and Other Zoonotic). 2018. Available online: (accessed on 13 November 2018).
- Sehrawat, S.; Suryawanshi, A.; Hirashima, M.; Rouse, B.T. Role of Tim-3/galectin-9 inhibitory interaction in viral-induced immunopathology: Shifting the balance toward regulators. J. Immunol. 2009, 182, 3191–3201.
- Merani, S.; Chen, W.; Elahi, S. The bitter side of sweet: The role of galectin-9 in immunopathogenesis of viral infections. Rev. Med. Virol. 2015, 25, 175–186.
More
