Clubroot is a major disease for the brassica crops, and it can not completely control by any other means except the resistant caultivars. MAS breeding helps us for the development of new resistant cultivars
- MAS
- Clubroot
- Resistant Genes:
1. Introduction
MAS is an indirect selection process where a trait of interest is selected based on a marker (morphological, biochemical, or DNA/RNA variation) linked to that trait. Selecting individuals with disease resistance using MAS involves identifying a marker allele that is linked to disease resistance rather than to the level of disease resistance. There are several types of DNA markers that have been used to identify disease resistance genes [1][2][3][4][5][6][7][8].
The complexity of plant–pathogen interaction is a problematic in the case of CR breeding due to the appearance of multiple races of the pathogen [9]. Combinations of different
CR genes exhibit higher resistance to the disease [10][11][12]. Though CR cultivars have been used widely for major production areas, field isolates of
P. brassicae
B. rapa
B. oleracea
P. brassicae. This suggests a serious risk that a resistance gene can be overcome by pathogen variants [13]. For example, seven CR canola cultivars were characterized for virulence in 106
P. brassicae
P. brassicae population overcame the resistance in at least one of the seven CR cultivars [14]. There are many reports that
CR
P. brassicae [15][16][17][18][19][10][20][21][22], but heterozygous
CR loci are less resistant than the homozygous state [23].
B. rapa
CR loci, which may confer differential (pathotype-specific) resistance to particular isolates of
P. brassicae, and sometimes this may have a large effect on resistance [9][15][24][25]. The NARO Institute of Vegetable and Tea Science (NIVTS) has developed a high CR Chinese cabbage cultivar, ‘Akimeki’, by the accumulation of
Crr1
Crr2
CRb
CR
P. brassicae
B. rapa
CR
CRa
CRk
CRc, were accumulated in Chinese cabbage through MAS [9] and the homozygous lines for the
CR
P. brassicae
CR genes could be controlled by the dose-dependent accumulation of CR proteins [26][27]. In
B. oleracea,
B. rapa and the level of resistance is low [28]. This might be due to the polygenic nature of resistance in
B. oleracea [11].
B. oleracea progeny were developed by accumulating major and minor QTLs to evaluate its effectiveness to the clubroot disease [29]. Three QTLs in the F
2
3 population from the cross between cabbage and kale line K269 were identified [10]. The accumulation of those three
CR genes showed broad resistance to three isolates. It was observed that only one major QTL PbBo(Anju)1 showed moderate resistance, whereas three minor QTLs without the major one showed distinct susceptibility [29]. Later, it was proven that PbBo(Anju)1 and three minor QTLs PbBo(Anju)2, PbBo(Anju)4, and PbBo(GC)1 play a critical role in the acquisition of resistance to clubroot disease [11][12]. Here,
PbBo(Anju)1
CR genes are also essential for achieving higher resistance [11][12]. Their effectiveness was verified for controlling disease involving various isolates of
P. brassicae [11]. Recently, two
CR
CRb
PbBa8.1
B. napus. The homozygous lines demonstrated a higher resistance than the heterozygous lines [30].
The Type A resistance to Fusarium wilt disease controlled by a single dominant gene has been successfully mapped and molecular markers have been developed: SSR marker KBrS003O1N10 [31], InDel markers M10 and A1 [6], Indel markers Bra012688m and Bra012689m [32][7], and DNA marker sets [8][33][34], which are used to generate a series of resistance cultivars (
Figure 1).
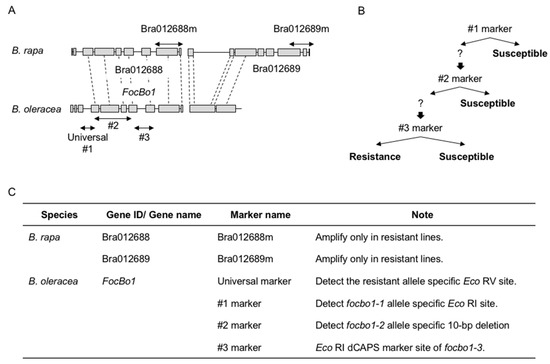
Figure 1. Schematic view of the alignment of resistance genes of Fusarium wilt disease. (A). DNA marker positions of resistance genes in
B. rapa and B. oleracea. Arrows indicate marker positions. (B). Scheme of marker assisted selection in B. oleracea. (C). DNA marker list for marker assisted selection in B. rapa and B. oleracea.
2. Influence and Results
Breeding cultivars that have resistance to both clubroot and Fusarium wilt is desired. However, inoculation tests against multiple pathogens or multiple races are difficult to perform on the same individual plant. Thus, DNA marker-based selection is useful for the identification of plants that have one Fusarium wilt resistance gene and multiple clubroot resistance genes. Furthermore, it is necessary to confirm whether these resistance genes are linked. In B. rapa, a Fusarium wilt resistance gene is located on chromosome 3, and
CRa/CRb
Crr3
CRk
CRa/CRb
Figure 2). Since recombination between these two genes can occur [7], it is possible to inherit both resistance genes. In
B. oleracea, a Fusarium wilt resistance gene is located on chromosome 7, and there is a minor QTL for clubroot resistance, PbBo(Anju)4, nearby this Fusarium wilt resistance gene. However, these loci are not completely linked to each other [8][32][35]. Therefore, it is possible to have both resistance genes. In
B. napus, the association between susceptibility to Fusarium wilt and clubroot resistance against pathotype 3 was found, and these two resistance genes are located about 10 cM apart [36]. However, recombination between these two genes has been reported [36], suggesting that it is possible to inherit both resistance genes and identify them by DNA marker-based selection.
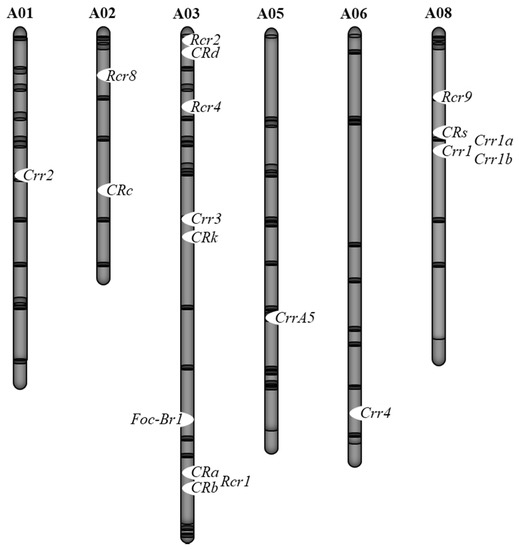
Figure 2. Chromosomal locations of clubroot resistant (CR) and Fusarium wilt resistant loci in B. rapa.
From the results from various researchers, it has been demonstrated that the DNA markers developed can select for the genes that are required for the acquisition of resistance, and these markers could be a powerful tool for resistance breeding in Brassica species. The novel breeding method developed can reinforce resistance by pyramiding
R
R
R
R
R
