Advances achieved with molecular biology and genomics technologies have permitted investigators to discover epigenetic mechanisms, such as DNA methylation and histone posttranslational modifications, which are critical for gene expression in almost all tissues and in brain health and disease. These advances have influenced much interest in understanding the dysregulation of epigenetic mechanisms in neurodegenerative disorders. Although these disorders diverge in their fundamental causes and pathophysiology, several involve the dysregulation of histone methylation-mediated gene expression. Interestingly, epigenetic remodeling via histone methylation in specific brain regions has been suggested to play a critical function in the neurobiology of psychiatric disorders, including that related to neurodegenerative diseases. Prominently, epigenetic dysregulation currently brings considerable interest as an essential player in neurodegenerative disorders, such as Alzheimer’s disease (AD), Parkinson’s disease (PD), Huntington’s disease (HD), Amyotrophic lateral sclerosis (ALS) and drugs of abuse, including alcohol abuse disorder, where it may facilitate connections between genetic and environmental risk factors or directly influence disease-specific pathological factors.
- epigenetics
- Alzheimer’s disease
- Parkinson’s disease
- Huntington’s disease
- Amyotrophic lateral sclerosis
- neuronal loss and alcohol
1. Introduction
Neurodegenerative (ND) disorders are among the leading bases of disability and death worldwide [1][2][3]. The neurodegeneration process involves progressive atrophy of neurons, leading to loss of neuronal connectivity and function followed by its demise, thereby adversely affecting brain function. Despite decades of basic and clinical research, most strategies designed to reverse degenerative brain diseases are analgesic. This is not surprising as neurodegeneration progresses quietly for decades before the appearance of symptoms. Most important advances in sequencing technologies have allowed the mapping of transcriptomic patterns in human postmortem brain tissues in various ND disorders, including in vitro and in vivo cell and animal models. These investigations facilitated the discovery of classical neurodegeneration pathways and uncovered novel targets, including synaptic degeneration in the majority of ND, that share several puzzling characteristics. For example, although large patient populations’ intense genetic evaluation has been performed, a substantial proportion of ND incidents have no known genetic origin [4], and only a few neuro-pathophysiology studies have identified gene mutations or defective genes. The majority of ND studies recognized the contribution of adverse environmental conditions, such as exposure to toxins, chemicals, nutritional deficits, social factors, drug abuse, and alcohol, leading to neurodegeneration and manifestation of pathology and behavioral defects. Many medications and therapies have been evaluated for these diseases, resulting in less than acceptable results [5][6]. Therefore, the necessity for novel treatments to improve symptoms and prevent ND progression is at an all-time high.
Neurodegenerative (ND) disorders are among the leading bases of disability and death worldwide [1,2,3]. The neurodegeneration process involves progressive atrophy of neurons, leading to loss of neuronal connectivity and function followed by its demise, thereby adversely affecting brain function. Despite decades of basic and clinical research, most strategies designed to reverse degenerative brain diseases are analgesic. This is not surprising as neurodegeneration progresses quietly for decades before the appearance of symptoms. Most important advances in sequencing technologies have allowed the mapping of transcriptomic patterns in human postmortem brain tissues in various ND disorders, including in vitro and in vivo cell and animal models. These investigations facilitated the discovery of classical neurodegeneration pathways and uncovered novel targets, including synaptic degeneration in the majority of ND, that share several puzzling characteristics. For example, although large patient populations’ intense genetic evaluation has been performed, a substantial proportion of ND incidents have no known genetic origin [4], and only a few neuro-pathophysiology studies have identified gene mutations or defective genes. The majority of ND studies recognized the contribution of adverse environmental conditions, such as exposure to toxins, chemicals, nutritional deficits, social factors, drug abuse, and alcohol, leading to neurodegeneration and manifestation of pathology and behavioral defects. Many medications and therapies have been evaluated for these diseases, resulting in less than acceptable results [5,6]. Therefore, the necessity for novel treatments to improve symptoms and prevent ND progression is at an all-time high.
2. PTM of DNA-Associated Histone Proteins
Histones are a highly conserved set of nuclear proteins that assemble in an octamer composed of pairs of H2A, H2B, H3, and H4. Nuclear DNA is highly condensed and wrapped around these nuclear proteins, nucleosomes, in the form of chromatin to accommodate chromosomes in the nuclei [7]. The histone proteins’ N-terminal tail domain undergoes chemical modification through PTMs of each specific amino acid residues, including acetylation, methylation, phosphorylation, and sumoylation [8]. This unique chemical PTM impacts the chromatin structure and recruitment of DNA binding factors and allows for the relaxing or compacting of the chromatin structure around particular gene loci, resulting in the activation or repression of specific gene expression [8]. Histone methylation is one of the PTM that regulates gene expression. Adding or removing methyl groups on lysine (K) residues of histones by histone methyltransferases (HMT) or histone demethylases (HMD) alters the structure of chromatin to facilitate (relaxed chromatin) or prevent (condensed chromatin) access of transcription factor proteins to genomic DNA, thereby guiding gene expression or repression in a more complex manner [9][10] (
Histones are a highly conserved set of nuclear proteins that assemble in an octamer composed of pairs of H2A, H2B, H3, and H4. Nuclear DNA is highly condensed and wrapped around these nuclear proteins, nucleosomes, in the form of chromatin to accommodate chromosomes in the nuclei [16]. The histone proteins’ N-terminal tail domain undergoes chemical modification through PTMs of each specific amino acid residues, including acetylation, methylation, phosphorylation, and sumoylation [17]. This unique chemical PTM impacts the chromatin structure and recruitment of DNA binding factors and allows for the relaxing or compacting of the chromatin structure around particular gene loci, resulting in the activation or repression of specific gene expression [17]. Histone methylation is one of the PTM that regulates gene expression. Adding or removing methyl groups on lysine (K) residues of histones by histone methyltransferases (HMT) or histone demethylases (HMD) alters the structure of chromatin to facilitate (relaxed chromatin) or prevent (condensed chromatin) access of transcription factor proteins to genomic DNA, thereby guiding gene expression or repression in a more complex manner [18,19] ().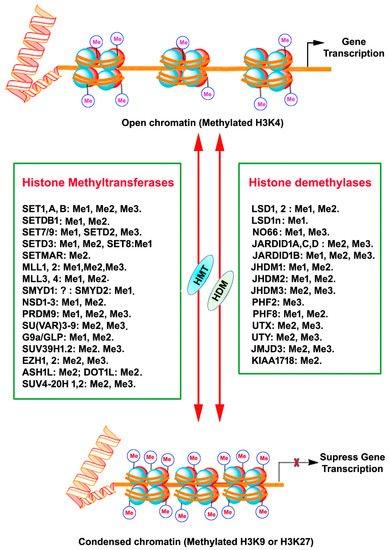
Figure 1.
3. Histone Methylation
Histone methylation usually occurs at the arginine or lysine N-terminal region, which leads to the activation or suppression of gene expression. Each arginine residue in histones can be subjected to monomethylation and symmetric/asymmetric dimethylation. Likewise, each lysine residue can be mono-, di- and tri-methylated, and also nearby residues can form different methylation combinations [11][12][13][14]. Methylation of all the arginine residue is catalyzed by the family of arginine methyltransferase (PRMT) enzymes. PRMT1 monomethylates and asymmetrically dimethylates arginine residues, thus resulting in the activation of gene expression. PRMT2 promotes gene suppression through monomethylation and symmetric dimethylation of arginine residues. Moreover, the cofactor-associated arginine methyltransferase (CARM1) promotes gene activation through monomethylation and asymmetric dimethylation of arginine residues. HMTs that catalyze the methylation of lysine amino acids of histone proteins and other proteins are called lysine methyltransferases (KMTs) [15] for their extensive enzymatic activity and precise substrate specificity (
Histone methylation usually occurs at the arginine or lysine N-terminal region, which leads to the activation or suppression of gene expression. Each arginine residue in histones can be subjected to monomethylation and symmetric/asymmetric dimethylation. Likewise, each lysine residue can be mono-, di- and tri-methylated, and also nearby residues can form different methylation combinations [20,21,22,23]. Methylation of all the arginine residue is catalyzed by the family of arginine methyltransferase (PRMT) enzymes. PRMT1 monomethylates and asymmetrically dimethylates arginine residues, thus resulting in the activation of gene expression. PRMT2 promotes gene suppression through monomethylation and symmetric dimethylation of arginine residues. Moreover, the cofactor-associated arginine methyltransferase (CARM1) promotes gene activation through monomethylation and asymmetric dimethylation of arginine residues. HMTs that catalyze the methylation of lysine amino acids of histone proteins and other proteins are called lysine methyltransferases (KMTs) [24] for their extensive enzymatic activity and precise substrate specificity (
Figure 1). All KMTs, except for KMT4, have a conserved Su (var)3–9, Enhancer of Zeste, Trithorax (SET) domain that presents catalytical activity [16]. The presence of a characteristic homologous sequence [15][17] has enabled the classification KMTs into well-defined subcategories. The KMTs that add the methyl group on H3K9, H3K27, and H4K20 are the main repressive KMTs, while the KMTs that add methyl group on H3K4, H3K14, and H3K36 are classified as activating KMTs [18]. One of the most highly investigated marks is H3 lysine (4) (H3K4) methylation, which is associated with active gene expression. Histone H3K4 methylation is catalyzed by a group (KMT2) of mixed lineage leukemia (MLL) proteins (SET1A, SET1B, MLL1, MLL2, MLL3, MLL4, and ASH1) [19][20][21]. H3K4me3 mark is found mainly in nucleosomes associated with the promoter regions of actively transcribed genes [22], while H3K4me2 is located in the gene bodies and enhancers associated with active genes [9]. H3K4me1 is found in enhancers, promoters, and at the 3′ end of active genes [23]. A family of methyltransferases (KMT1) (SUV39H1, SUV39H2, G9A/GLP and ESET/SETDB1; KMT8: RIZ1) catalyze histone H3K9 methylation and catalytical activity differ with catalyzing substrates and resulting products. Histone H3K9 trimethylation (H3K9me3) is catalyzed by SUV39 and results in a heterochromatin structure and transcriptional suppression [24]. Histone H3K9 dimethylation (H3K9me2) is catalyzed by G9A and results in a euchromatin structure and suppressed gene expression [25][26]. G9A-like protein (GLP) forms a hetero polypeptide complex with G9A, and, collectively, they catalyze the H3K9 dimethylation [27]. ESET/SETDB1 catalyzes the trimethylation of H3K9 [28], which results in the inhibition of gene expression. Among the alterations linked with transcription elongation, histone H3K36 trimethylation modification occurring at nucleosomes in the 3′ region of the transcription region of active genes [29]. H3K36me3 is catalyzed by KMT3 (SET2, SYMD2 and NSD1) enzymes [30][31][32]. Histone H3K79 di- and tri-methylation are catalyzed by DOT1L enzymes (KMT4) and are associated with gene expression activation [33]. Histone H4K20 mono- or tri-methylation is catalyzed by SET8 and SUV420H1/2 enzymes (KMT5), respectively, associated with gene inactivation [34]. The monomethylation of H3K27 often increases the expression of target genes, whereas the addition of three methyl groups at the same site usually suppresses gene transcription [35]. Histone H3K27me1 is catalyzed by ATXR5 and ATXR6 or TXR1 [36][37] and H3K27me3 is established by Enhancer of Zeste 2 (EZH2) of the polycomb repressive complex 2 (PRC2) [38][39][40].
). All KMTs, except for KMT4, have a conserved Su (var)3–9, Enhancer of Zeste, Trithorax (SET) domain that presents catalytical activity [25]. The presence of a characteristic homologous sequence [24,26] has enabled the classification KMTs into well-defined subcategories. The KMTs that add the methyl group on H3K9, H3K27, and H4K20 are the main repressive KMTs, while the KMTs that add methyl group on H3K4, H3K14, and H3K36 are classified as activating KMTs [27]. One of the most highly investigated marks is H3 lysine (4) (H3K4) methylation, which is associated with active gene expression. Histone H3K4 methylation is catalyzed by a group (KMT2) of mixed lineage leukemia (MLL) proteins (SET1A, SET1B, MLL1, MLL2, MLL3, MLL4, and ASH1) [28,29,30]. H3K4me3 mark is found mainly in nucleosomes associated with the promoter regions of actively transcribed genes [31], while H3K4me2 is located in the gene bodies and enhancers associated with active genes [18]. H3K4me1 is found in enhancers, promoters, and at the 3′ end of active genes [32]. A family of methyltransferases (KMT1) (SUV39H1, SUV39H2, G9A/GLP and ESET/SETDB1; KMT8: RIZ1) catalyze histone H3K9 methylation and catalytical activity differ with catalyzing substrates and resulting products. Histone H3K9 trimethylation (H3K9me3) is catalyzed by SUV39 and results in a heterochromatin structure and transcriptional suppression [33]. Histone H3K9 dimethylation (H3K9me2) is catalyzed by G9A and results in a euchromatin structure and suppressed gene expression [34,35]. G9A-like protein (GLP) forms a hetero polypeptide complex with G9A, and, collectively, they catalyze the H3K9 dimethylation [36]. ESET/SETDB1 catalyzes the trimethylation of H3K9 [37], which results in the inhibition of gene expression. Among the alterations linked with transcription elongation, histone H3K36 trimethylation modification occurring at nucleosomes in the 3′ region of the transcription region of active genes [38]. H3K36me3 is catalyzed by KMT3 (SET2, SYMD2 and NSD1) enzymes [39,40,41]. Histone H3K79 di- and tri-methylation are catalyzed by DOT1L enzymes (KMT4) and are associated with gene expression activation [42]. Histone H4K20 mono- or tri-methylation is catalyzed by SET8 and SUV420H1/2 enzymes (KMT5), respectively, associated with gene inactivation [43]. The monomethylation of H3K27 often increases the expression of target genes, whereas the addition of three methyl groups at the same site usually suppresses gene transcription [44]. Histone H3K27me1 is catalyzed by ATXR5 and ATXR6 or TXR1 [45,46] and H3K27me3 is established by Enhancer of Zeste 2 (EZH2) of the polycomb repressive complex 2 (PRC2) [47,48,49].
4. Alzheimer’s Disease (AD)
AD is the most established neurodegenerative disorder in the world. Emerging findings indicate that epigenetic dysregulation of gene expression may play a significant role in aging and ND [41][42][43][44]. Although global accumulation or loss of histone methylation proteins and the subsequent alteration of the expression of many genes are shown in aging and cognitive functioning [23], the role of the diverse array of histone methylation, even in AD, has not been identified [41][45]. Additionally, a clinical study has shown modifications in H2B K108 and H4 arginine (R) 55 methylation in the frontal cortex from human donors with AD [46], suggesting that histone methylation may be a new potential therapeutic target to treat AD-related cognitive abnormalities. A postmortem AD brain study reported elevated levels of H3K9me2 proteins in the occipital cortex compared to nondemented and age-matched controls [47]. The inhibition of G9a/GLP catalytic activity by BIX 01294 prevents the Aβ oligomer-induced late-LTP and synaptic tagging and capture (STC) deficits by releasing the transcription repression of the Bdnf gene [48]. Moreover, BIX 01294 treatment in the hippocampus slices rescued Aβ oligomer-induced suppression of Bdnf gene expression [48]. Similarly, another study [49] has shown significantly elevated H3K9me2 and Emt1 (G9a) and Emt2 (GLP) in the PFC and HP from late-stage FAD mice and human patients with AD. These mice also exhibited reduced glutamate receptor transcription and many AD-like cognitive deficits. The elevated global H3K9me2 in FAD mice was also significantly enriched at the transcription start site regions of glutamate receptors (Gria2/GluA2 and Grin2b/NR2B genes), indicating that the loss of glutamate receptor transcription in AD is due to aberrant histone H3K9 dimethylation. Interestingly, pharmacological inhibition of H3K9me2 formation by GLP/G9a improved Gria2/GluA2, Grin2b/NR2B genes’ transcription, and that of additional genes (e.g., SHANK2) that are implicated in AD. These studies suggest that pharmacological inhibition (BIX 01294) of GLP/G9a normalizes multiple target genes and restores synaptic function and cognitive functioning in aged FAD mice [49]. Consistent with the above findings, exposure of mature human cortical neurons, derived from human embryonic stem cells, to Aβ significantly enhanced G9a, and inhibited AMPAR-mediated whole-cell current and excitatory postsynaptic current (EPSC) [50]. Further, the addition of BIX 01294 rescued EPSC in Aβ exposed human cortical neurons [50]. Interestingly, AD pathological hallmarks (hyperphosphorylated tau and Aβ plaques and cognitive deficits) exhibited by children and young adults in polluted cities showed reduced H3K9me2/me3 in postmortem prefrontal white matter [51]. Another more specific G9a/GLP inhibitor (UNC0642) was used to rescue 5XFAD cognition impairment and the H3K9me2 levels in the hippocampus [52]. The UNC0642 was successful in improving gene expression (Nuclear Factor erythroid-2-Related Factor 2 (NRF2), Heme oxygenase decycling 1 (Hmox1), Nerve growth factor (Ngf), Nerve growth factor inducible (Vgf), BDNF, and Synaptophysin (SYN)), protecting pathological changes (Reactive Oxygen Species, ROS; neuroinflammatory markers, such as Interleukin 6 (Il-6), Tumor necrosis factor-alpha (Tnf-α) gene expression, and Glial fibrillary acidic protein (GFAP), reduction in β-amyloid plaques) in 5XFAD mice [52]. A recent genetic study [53] of aging human brains has suggested that the strongly linked module with cognitive deficits is enriched with genes that regulate chromatin remodeling [53]. In contrast to the above-discussed findings, reduced H3K9me2 levels in the CA1 region of middle-aged and AD stages I-VI [54] were observed. A positive correlation between H3K4me3 marks and the long noncoding RNAs’ (lncRNA) gene expression level and a negative association between H3K27me3 marks and the lncRNA gene expression level was found in the CK-p25 AD model [55]. In contrast, decreased H3K4me3 with no change in H3K27me3 marks at the ANK1 gene locus in postmortem AD brains was reported [56]. Interestingly, an age-related increase in H3K27me3 was observed in neurofilament (NF)-labeled calretinin-positive interneurons [57]. However, amyloid plaque deposition and its sequelae failed to alter global H3K27me3 in NF-positive calretinin-labeled interneurons. The mislocalization of H3K4me3 between the nucleus vs. cytoplasm was observed in the medial temporal gyrus of the human postmortem AD brain [58]. The mislocalized cytoplasmic H3K4me3 was associated with pre-tangles and NFTs in late AD brains [58]. Similarly, a loss of nuclear H3K4me3 mark in hippocampal subregions but not cytoplasmic H3K4me3 mark was found in an AD animal (3×Tg) model, which develops plaques and NFTs [58]. Similarly, the H3K4m3 mark, as well as specific HMTs, such as KMT2A-D in the PFC nuclear fraction from human AD, was significantly enhanced without affecting H3K27me3 or H3K4me marks [59]. A similar increase in H3K4me3 and Kmt2a was also found in P301S transgenic Tau mice (PS19) [59]. These epigenetic advances provide robust experimental evidence using multiple AD models and suggest that restoring histone methylation’s homeostasis specifically mediated by G9a/GLP and KMT2A-D may perhaps be a potential therapeutic strategy to treat AD-related neurodegenerative disorders. Given the multifactorial characteristics of AD, targeting a single or a couple of abnormal genes is unlikely to prevent pathophysiological and behavioral abnormalities. Well-characterized histone methylation targeted pharmacotherapy in the future may offer the advantages of having broad, multifunctional actions and being able to target a network of genes essential for neuronal survival and function.
AD is the most established neurodegenerative disorder in the world. Emerging findings indicate that epigenetic dysregulation of gene expression may play a significant role in aging and ND [86,87,88,89]. Although global accumulation or loss of histone methylation proteins and the subsequent alteration of the expression of many genes are shown in aging and cognitive functioning [32], the role of the diverse array of histone methylation, even in AD, has not been identified [86,90]. Additionally, a clinical study has shown modifications in H2B K108 and H4 arginine (R) 55 methylation in the frontal cortex from human donors with AD [91], suggesting that histone methylation may be a new potential therapeutic target to treat AD-related cognitive abnormalities. A postmortem AD brain study reported elevated levels of H3K9me2 proteins in the occipital cortex compared to nondemented and age-matched controls [92]. The inhibition of G9a/GLP catalytic activity by BIX 01294 prevents the Aβ oligomer-induced late-LTP and synaptic tagging and capture (STC) deficits by releasing the transcription repression of the Bdnf gene [93]. Moreover, BIX 01294 treatment in the hippocampus slices rescued Aβ oligomer-induced suppression of Bdnf gene expression [93]. Similarly, another study [94] has shown significantly elevated H3K9me2 and Emt1 (G9a) and Emt2 (GLP) in the PFC and HP from late-stage FAD mice and human patients with AD. These mice also exhibited reduced glutamate receptor transcription and many AD-like cognitive deficits. The elevated global H3K9me2 in FAD mice was also significantly enriched at the transcription start site regions of glutamate receptors (Gria2/GluA2 and Grin2b/NR2B genes), indicating that the loss of glutamate receptor transcription in AD is due to aberrant histone H3K9 dimethylation. Interestingly, pharmacological inhibition of H3K9me2 formation by GLP/G9a improved Gria2/GluA2, Grin2b/NR2B genes’ transcription, and that of additional genes (e.g., SHANK2) that are implicated in AD. These studies suggest that pharmacological inhibition (BIX 01294) of GLP/G9a normalizes multiple target genes and restores synaptic function and cognitive functioning in aged FAD mice [94]. Consistent with the above findings, exposure of mature human cortical neurons, derived from human embryonic stem cells, to Aβ significantly enhanced G9a, and inhibited AMPAR-mediated whole-cell current and excitatory postsynaptic current (EPSC) [95]. Further, the addition of BIX 01294 rescued EPSC in Aβ exposed human cortical neurons [95]. Interestingly, AD pathological hallmarks (hyperphosphorylated tau and Aβ plaques and cognitive deficits) exhibited by children and young adults in polluted cities showed reduced H3K9me2/me3 in postmortem prefrontal white matter [96]. Another more specific G9a/GLP inhibitor (UNC0642) was used to rescue 5XFAD cognition impairment and the H3K9me2 levels in the hippocampus [97]. The UNC0642 was successful in improving gene expression (Nuclear Factor erythroid-2-Related Factor 2 (NRF2), Heme oxygenase decycling 1 (Hmox1), Nerve growth factor (Ngf), Nerve growth factor inducible (Vgf), BDNF, and Synaptophysin (SYN)), protecting pathological changes (Reactive Oxygen Species, ROS; neuroinflammatory markers, such as Interleukin 6 (Il-6), Tumor necrosis factor-alpha (Tnf-α) gene expression, and Glial fibrillary acidic protein (GFAP), reduction in β-amyloid plaques) in 5XFAD mice [97]. A recent genetic study [98] of aging human brains has suggested that the strongly linked module with cognitive deficits is enriched with genes that regulate chromatin remodeling [98]. In contrast to the above-discussed findings, reduced H3K9me2 levels in the CA1 region of middle-aged and AD stages I-VI [99] were observed. A positive correlation between H3K4me3 marks and the long noncoding RNAs’ (lncRNA) gene expression level and a negative association between H3K27me3 marks and the lncRNA gene expression level was found in the CK-p25 AD model [100]. In contrast, decreased H3K4me3 with no change in H3K27me3 marks at the ANK1 gene locus in postmortem AD brains was reported [101]. Interestingly, an age-related increase in H3K27me3 was observed in neurofilament (NF)-labeled calretinin-positive interneurons [102]. However, amyloid plaque deposition and its sequelae failed to alter global H3K27me3 in NF-positive calretinin-labeled interneurons. The mislocalization of H3K4me3 between the nucleus vs. cytoplasm was observed in the medial temporal gyrus of the human postmortem AD brain [103]. The mislocalized cytoplasmic H3K4me3 was associated with pre-tangles and NFTs in late AD brains [103]. Similarly, a loss of nuclear H3K4me3 mark in hippocampal subregions but not cytoplasmic H3K4me3 mark was found in an AD animal (3×Tg) model, which develops plaques and NFTs [103]. Similarly, the H3K4m3 mark, as well as specific HMTs, such as KMT2A-D in the PFC nuclear fraction from human AD, was significantly enhanced without affecting H3K27me3 or H3K4me marks [104]. A similar increase in H3K4me3 and Kmt2a was also found in P301S transgenic Tau mice (PS19) [104]. These epigenetic advances provide robust experimental evidence using multiple AD models and suggest that restoring histone methylation’s homeostasis specifically mediated by G9a/GLP and KMT2A-D may perhaps be a potential therapeutic strategy to treat AD-related neurodegenerative disorders. Given the multifactorial characteristics of AD, targeting a single or a couple of abnormal genes is unlikely to prevent pathophysiological and behavioral abnormalities. Well-characterized histone methylation targeted pharmacotherapy in the future may offer the advantages of having broad, multifunctional actions and being able to target a network of genes essential for neuronal survival and function.5. Huntington’s Disease (HD)
HD is a late-onset, autosomal progressive neurodegenerative disorder caused by the trinucleotide CAG repeat in the coding for glutamine (Q) in exon 1 of the Huntingtin (Htt) gene [60], leading to the motor, cognitive and psychiatric symptomatology [61][62]. Modification of the chromatin structure and deregulation of neuronal gene transcription are prominent features associated with the earliest stage of HD. Studies in recent decades using patients and multiple animal models of HD have identified histone modifications (acetylation, methylation, ubiquitylation, and phosphorylation) (see review [63]) and DNA modifications as significant epigenetic modifications that regulate gene expression in HD. The pharmacological approaches directed to correct some of those epigenetic changes have offered potential in treating HD (see review [64]). The following discussion presents the most recent advances in histone methylation-mediated epigenetics for potential HD disease interventions.
HD is a late-onset, autosomal progressive neurodegenerative disorder caused by the trinucleotide CAG repeat in the coding for glutamine (Q) in exon 1 of the Huntingtin (Htt) gene [105], leading to the motor, cognitive and psychiatric symptomatology [106,107]. Modification of the chromatin structure and deregulation of neuronal gene transcription are prominent features associated with the earliest stage of HD. Studies in recent decades using patients and multiple animal models of HD have identified histone modifications (acetylation, methylation, ubiquitylation, and phosphorylation) (see review [108]) and DNA modifications as significant epigenetic modifications that regulate gene expression in HD. The pharmacological approaches directed to correct some of those epigenetic changes have offered potential in treating HD (see review [109]). The following discussion presents the most recent advances in histone methylation-mediated epigenetics for potential HD disease interventions.Enhanced H3K9me2 has been found in murine models of HD [65][66][67], suggesting that a gain of specific HMT or loss of HDM function appears to be associated with the pathogenic gene suppression observed in disease progression. SETDB1, a methyltransferase that explicitly targets the H3K9me3 [68], was enhanced in patients with HD and transgenic R6/2 HD mice [69]. Based on these observations, it was evident that neuronal levels of SETDB1 and H3K9me3 might be an indication of nucleosomal dysfunction in HD [70][71]. Similarly, monoallelic deletion of cyclic AMP response element-binding (CREB) protein (CBP) causes enhanced SETDB1 gene expression and H3K9 hypermethylation [72]. More importantly, enhanced H3K9me3 was found to promote the establishment of large constitutive heterochromatin domains and has been shown to promote both global and local [72][73] suppression of gene expression, including enrichment at the muscarinic acetylcholine (Ach) receptor 1 (CHRM1) gene promoter in HD striatal cells [72]. Consistent with these findings, the combined use of mithramycin, a clinically approved guanosine–cytosine-rich DNA binding antitumor antibiotic, and cystamine suppressed ESET expression by reducing H3K9 hypermethylation and significantly rescued behavioral and neuropathological phenotypes and extended survival by over 40% in R6/2 HD mice [72]. These observations suggest that epigenome regulation via H3K9me3 is vital for neuronal survival in HD and appears to be a potential treatment target in patients with HD.
Enhanced H3K9me2 has been found in murine models of HD [110,111,112], suggesting that a gain of specific HMT or loss of HDM function appears to be associated with the pathogenic gene suppression observed in disease progression. SETDB1, a methyltransferase that explicitly targets the H3K9me3 [113], was enhanced in patients with HD and transgenic R6/2 HD mice [114]. Based on these observations, it was evident that neuronal levels of SETDB1 and H3K9me3 might be an indication of nucleosomal dysfunction in HD [115,116]. Similarly, monoallelic deletion of cyclic AMP response element-binding (CREB) protein (CBP) causes enhanced SETDB1 gene expression and H3K9 hypermethylation [117]. More importantly, enhanced H3K9me3 was found to promote the establishment of large constitutive heterochromatin domains and has been shown to promote both global and local [117,118] suppression of gene expression, including enrichment at the muscarinic acetylcholine (Ach) receptor 1 (CHRM1) gene promoter in HD striatal cells [117]. Consistent with these findings, the combined use of mithramycin, a clinically approved guanosine–cytosine-rich DNA binding antitumor antibiotic, and cystamine suppressed ESET expression by reducing H3K9 hypermethylation and significantly rescued behavioral and neuropathological phenotypes and extended survival by over 40% in R6/2 HD mice [117]. These observations suggest that epigenome regulation via H3K9me3 is vital for neuronal survival in HD and appears to be a potential treatment target in patients with HD.Besides, studies have also shown a functional interaction of the Htt gene with protein arginine methyltransferase 5 (PRMT5), an enzyme mediating the dimethylation of arginine (R) of essential cellular proteins, including histones and spliceosomal proteins [74]. Htt has been shown to activate PRMT5 and reduce arginine dimethylation of histone H2A and H4 in primary cultured neurons and HD brains [74]. Consistent with this observation, the expression of PRMT5/methylosome protein 50 (MEP50) complexes, or the genetic ablation of the JMJD6 (H4R3Me2 demethylase), rescued the toxic effects of mutant Htt in primary cortical neurons [74], indicating that PRMT5 loss may be responsible, at least in part, for HD pathogenesis. It was shown that Htt null embryos exhibited impaired PRC2 [75], a methyltransferase complex containing Ezh2 that adds the trimethyl group to H3K27 and maintains the gene expression pattern by regulating the chromatin structure in mitotic and postmitotic cells in the brain [76][77]. Similarly, the lack of Htt in embryos reduced histone H3K27me3 and impaired homeobox (Hox) gene expression and trophoblast giant cell differentiation, causing paternal X chromosome inactivation [75]. Consistent with the results, genome-wide analysis in
Besides, studies have also shown a functional interaction of the Htt gene with protein arginine methyltransferase 5 (PRMT5), an enzyme mediating the dimethylation of arginine (R) of essential cellular proteins, including histones and spliceosomal proteins [119]. Htt has been shown to activate PRMT5 and reduce arginine dimethylation of histone H2A and H4 in primary cultured neurons and HD brains [119]. Consistent with this observation, the expression of PRMT5/methylosome protein 50 (MEP50) complexes, or the genetic ablation of the JMJD6 (H4R3Me2 demethylase), rescued the toxic effects of mutant Htt in primary cortical neurons [119], indicating that PRMT5 loss may be responsible, at least in part, for HD pathogenesis. It was shown that Htt null embryos exhibited impaired PRC2 [120], a methyltransferase complex containing Ezh2 that adds the trimethyl group to H3K27 and maintains the gene expression pattern by regulating the chromatin structure in mitotic and postmitotic cells in the brain [121,122]. Similarly, the lack of Htt in embryos reduced histone H3K27me3 and impaired homeobox (Hox) gene expression and trophoblast giant cell differentiation, causing paternal X chromosome inactivation [120]. Consistent with the results, genome-wide analysis inHtt
WT and targeted inactivation of both copies ofHtt (dKO) genotypes suggested that the loss of Htt triggered a significant reduction in the total number of H3K27me3-marked promoters [78]. These findings significantly imply that huntingtin is required to retain the H3K27 trimethyl group added by PRC2 efficiently. The effect of huntingtin on facilitating the enrichment of H3K27me3 could be achieved through the physical interaction between full-length huntingtin and the PRC2 complex [75], such as the Ezh2 or Suz12 or Utx (demethylates H3K27me3) [79][80].
(dKO) genotypes suggested that the loss of Htt triggered a significant reduction in the total number of H3K27me3-marked promoters [123]. These findings significantly imply that huntingtin is required to retain the H3K27 trimethyl group added by PRC2 efficiently. The effect of huntingtin on facilitating the enrichment of H3K27me3 could be achieved through the physical interaction between full-length huntingtin and the PRC2 complex [120], such as the Ezh2 or Suz12 or Utx (demethylates H3K27me3) [68,124].The additional function of PRC2 in HD was supported by H3K27me3 ChIP-sequencing in neuronal chromatin obtained from the HD postmortem PFC and non-neurologic controls. The findings suggest the loss of neuronal PRC2-H3K27me3 sites and the upregulation of some PRC2 target genes associated mainly with Hox gene clusters and developmentally related proteins in the HD-affected human brain [81][82][83][84]. Remarkably, loss of PRC2 levels in adult neurons was associated with the derepression of selected PRC2 target genes, followed by loss of neuronal functions and survival, therefore further strengthening the observation that persistent dysregulation of PRC2, in addition to other H3K27me3-controlling enzymes, may cause systemic neurodegeneration in HD [85]. Nevertheless, the presence of an expanded CAG tract is not only related to changes in the PRC2 pattern and changes in histone H3K27me3 enrichment, causing reduced RNA expression [78]. This observation emphasizes that histone-modifying enzymes and chromatin remodeling factors do not function as a single molecule but act as a supermolecular complex to control gene transcription in a coordinated but complementary manner (repression and activation) [86].
The additional function of PRC2 in HD was supported by H3K27me3 ChIP-sequencing in neuronal chromatin obtained from the HD postmortem PFC and non-neurologic controls. The findings suggest the loss of neuronal PRC2-H3K27me3 sites and the upregulation of some PRC2 target genes associated mainly with Hox gene clusters and developmentally related proteins in the HD-affected human brain [125,126,127,128]. Remarkably, loss of PRC2 levels in adult neurons was associated with the derepression of selected PRC2 target genes, followed by loss of neuronal functions and survival, therefore further strengthening the observation that persistent dysregulation of PRC2, in addition to other H3K27me3-controlling enzymes, may cause systemic neurodegeneration in HD [129]. Nevertheless, the presence of an expanded CAG tract is not only related to changes in the PRC2 pattern and changes in histone H3K27me3 enrichment, causing reduced RNA expression [123]. This observation emphasizes that histone-modifying enzymes and chromatin remodeling factors do not function as a single molecule but act as a supermolecular complex to control gene transcription in a coordinated but complementary manner (repression and activation) [130].Notably, the diminished enrichment of H3K4me3, a mark of active gene transcription, has been associated with impaired transcription of target genes (Bdnf, Penk1, Drd2) in both human HD postmortem brains as well as the cortex and striatum of R6/2 mice [87]. Reduced H3K4me3 enrichment was observed as the RE-1 silencing transcription factor/neuron-restrictive silencer factor (REST/NRSF) promoter II [87], therefore indicating that impaired transcription might be a result of changes in chromatin structure at the REST binding site on the
Notably, the diminished enrichment of H3K4me3, a mark of active gene transcription, has been associated with impaired transcription of target genes (Bdnf, Penk1, Drd2) in both human HD postmortem brains as well as the cortex and striatum of R6/2 mice [131]. Reduced H3K4me3 enrichment was observed as the RE-1 silencing transcription factor/neuron-restrictive silencer factor (REST/NRSF) promoter II [131], therefore indicating that impaired transcription might be a result of changes in chromatin structure at the REST binding site on theBDNF gene locus [87]. These observations are in line with previous results showing changed REST and BDNF signaling in HD [88][89]. Furthermore, the knockdown of JARID1C by ShRNA (demethylates H3K4me3) caused the upregulation of Bdnf gene expression in primary cortical neurons derived from the BACHD mouse HD model and improved the health of these neurons, indicating neuroprotection against HD in this model. Similarly, enhanced JARID1C was also observed in HD
gene locus [131]. These observations are in line with previous results showing changed REST and BDNF signaling in HD [132,133]. Furthermore, the knockdown of JARID1C by ShRNA (demethylates H3K4me3) caused the upregulation of Bdnf gene expression in primary cortical neurons derived from the BACHD mouse HD model and improved the health of these neurons, indicating neuroprotection against HD in this model. Similarly, enhanced JARID1C was also observed in HDHtt (Q150) knock-in mice [90]. Notably, JARID1C, which has been shown to interact with REST [91]), and the NRSE motif are strongly enriched near sites of reduced H3K4me3 in R6/7 mice [87]. A similarly reduced function of H3K4me3 at the genome-wide level in HD postmortem PFC tissues [81][92] was reported. These findings indicate that chromatin-remodeling enzymes, including SETDB1, PRMT5, Ezh2, JARID1C, may be potential therapeutic targets for HD treatment [87], further strengthening the observation that repressive neuronal chromatin mediated by histone methylation may have a crucial role in HD pathophysiology, including ND.
(Q150) knock-in mice [134]. Notably, JARID1C, which has been shown to interact with REST [65]), and the NRSE motif are strongly enriched near sites of reduced H3K4me3 in R6/7 mice [131]. A similarly reduced function of H3K4me3 at the genome-wide level in HD postmortem PFC tissues [125,135] was reported. These findings indicate that chromatin-remodeling enzymes, including SETDB1, PRMT5, Ezh2, JARID1C, may be potential therapeutic targets for HD treatment [131], further strengthening the observation that repressive neuronal chromatin mediated by histone methylation may have a crucial role in HD pathophysiology, including ND.6. Parkinson’s Disease (PD)
PD is the second most predominant neurodegenerative disorder in the world after AD [93]. The most significant pathology of PD is the demise of dopaminergic neurons in the substantia nigra with Lewy bodies (aggregated alpha-synuclein and ubiquitin-protein and damaged nerve cells as cytoplasmic inclusions) [94][95][96]. PD is characterized by motor symptoms (bradykinesia, tremor, postural instability, and rigidity) and nonmotor symptoms (cognitive issues, autonomic dysfunction, and REM sleep behavior disorder) [94]. However, the mechanisms involved in PD pathogenesis have not been fully identified. The specific loss of dopaminergic neurons in PD has been believed to be a consequence of complex interactions between genetic and environmental factors. Nevertheless, the characteristics of the relationship between the two significant changes remain to be established. A growing body of studies have begun to support epigenetic events, such as DNA methylation and histone modification in PD progression [97][98][99].
PD is the second most predominant neurodegenerative disorder in the world after AD [136]. The most significant pathology of PD is the demise of dopaminergic neurons in the substantia nigra with Lewy bodies (aggregated alpha-synuclein and ubiquitin-protein and damaged nerve cells as cytoplasmic inclusions) [137,138,139]. PD is characterized by motor symptoms (bradykinesia, tremor, postural instability, and rigidity) and nonmotor symptoms (cognitive issues, autonomic dysfunction, and REM sleep behavior disorder) [137]. However, the mechanisms involved in PD pathogenesis have not been fully identified. The specific loss of dopaminergic neurons in PD has been believed to be a consequence of complex interactions between genetic and environmental factors. Nevertheless, the characteristics of the relationship between the two significant changes remain to be established. A growing body of studies have begun to support epigenetic events, such as DNA methylation and histone modification in PD progression [140,141,142].
A mounting number of studies overwhelmingly suggest the critical function of the histone modification and PD-associated α-synuclein coding gene SNCA expression [100], meaning that histone methylation also may have a crucial role in the regulation of the SNCA gene. Interestingly, overexpression of α-SYN in flies and neuronal cells, such as SH-SY5Y enhanced G9a, H3K9me1, and H3K9me2 levels and H3K9me2-target genes (L1cam, Snap25) eventually lead to impaired synaptic activity [101]. In contrast, H3K4me3 was significantly enriched at the
A mounting number of studies overwhelmingly suggest the critical function of the histone modification and PD-associated α-synuclein coding gene SNCA expression [143], meaning that histone methylation also may have a crucial role in the regulation of the SNCA gene. Interestingly, overexpression of α-SYN in flies and neuronal cells, such as SH-SY5Y enhanced G9a, H3K9me1, and H3K9me2 levels and H3K9me2-target genes (L1cam, Snap25) eventually lead to impaired synaptic activity [144]. In contrast, H3K4me3 was significantly enriched at the
SNCA promoter region in postmortem brain samples from patients with PD and matched controls [102]. Similarly, using dead Cas9-Suntag system-mediated locus-specific approaches, the reduction in H3K4me3 from the
promoter region in postmortem brain samples from patients with PD and matched controls [145]. Similarly, using dead Cas9-Suntag system-mediated locus-specific approaches, the reduction in H3K4me3 from the
SNCA promoter reduced α-synuclein levels in neuronal cell lines and PD-derived induced pluripotent stem cell lines (iPSCs) [102]. Significant H3K4me3 enrichment was observed at the
promoter reduced α-synuclein levels in neuronal cell lines and PD-derived induced pluripotent stem cell lines (iPSCs) [145]. Significant H3K4me3 enrichment was observed at the
SNCA promoter in the neuronal nuclei (NeuN) of positive neurons of substantia nigra (SN) tissue samples from PD patients. Interestingly, significant reductions in H3K4me3 and H3K27me3 marks were observed in SH–SY5Y cells treated with the neurotoxin 6-hydroxydopamine (6-OHDA) [103]. In the same model, pretreatment with GSK-J4, a potent inhibitor of KDM6A/B and KDM5B/C (demethylates H3K27me3/me2 and H3K4me3/me2 respectively) [104][105], significantly prevented H3K4me3 and H3K27me3 marks’ decrease [103]. In contrast to the cell culture model, increased H3K27me3 levels were found in PD patients’ brains [102], indicating the possible role of PRC2 in vivo PD models. These findings indicate that abnormal histone methylation, such as H3K4me3 (gene activation) and H3K27me3 (gene suppression), control gene expression (e.g., α-synuclein) in SN neurons from patients with PD. Because specific histone methylation is one of the central regulators of gene expression, future investigation is warranted to understand further how specific histone methylation of the gene s, including the
promoter in the neuronal nuclei (NeuN) of positive neurons of substantia nigra (SN) tissue samples from PD patients. Interestingly, significant reductions in H3K4me3 and H3K27me3 marks were observed in SH–SY5Y cells treated with the neurotoxin 6-hydroxydopamine (6-OHDA) [146]. In the same model, pretreatment with GSK-J4, a potent inhibitor of KDM6A/B and KDM5B/C (demethylates H3K27me3/me2 and H3K4me3/me2 respectively) [147,148], significantly prevented H3K4me3 and H3K27me3 marks’ decrease [146]. In contrast to the cell culture model, increased H3K27me3 levels were found in PD patients’ brains [145], indicating the possible role of PRC2 in vivo PD models. These findings indicate that abnormal histone methylation, such as H3K4me3 (gene activation) and H3K27me3 (gene suppression), control gene expression (e.g., α-synuclein) in SN neurons from patients with PD. Because specific histone methylation is one of the central regulators of gene expression, future investigation is warranted to understand further how specific histone methylation of the gene s, including the
SNCA
gene, directly regulates the accessibility of transcription factors that can access gene regulatory regions. Finally, there are still undiscovered histone methylation marks that may regulate synaptic and pathological abnormalities found in PD. Those may also have a considerable impact on regulating chromatin structure and overall effects on gene transcription in PD and should be of future interest.
