In this entry, new pyrimidine derivatives were designed, synthesized and analyzed in terms of their anticancer properties. The tested compounds were evaluated in vitro for their antitumor activity. The cytotoxic effect on normal human dermal fibroblasts (NHDF) was also determined. According to the results, all the tested compounds exhibited inhibitory activity on the proliferation of all lines of cancer cells (colon adenocarcinoma (LoVo), resistant colon adenocarcinoma (LoVo/DX), breast cancer (MCF-7), lung cancer (A549), cervical cancer (HeLa), human leukemic lymphoblasts (CCRF-CEM) and human monocytic (THP-1)).
- pyrimidine
- anticancer
- 3
- 4-dihydronaphthalen
- 6-hydrazinopyrimidine
- lipophilicity
- QSAR study
- topoisomerase II
- DNA intercalating
1. Introduction
As reported by the WHO, 18 million people are presently living with cancer; over 9 million people died from cancer in 2018. Due to the lack of effective and selectively acting anticancer therapies, these numbers are still increasing. The compounds based on the scaffold of pyrimidine exhibit a broad spectrum of pharmacological activity such as anti-inflammatory [1], antimicrobial [2][3], anti-HIV [2], antidiabetic [4] and anticancer activity [5][6][7]. The most recognized drugs based on analogs of pyrimidines are antibacterial (sulfadiazine, trimethoprim), antiviral (trifluridine, idoxuridine), anti-malarial (sulfadoxine), anti-HIV (Retrovir (zidovudine), stavudine), anti-tuberculosis (viomycin), anticancer (5-fluorouracil) agents. The data also show that the adjunction of a [(dialkylamino)alkyl]amino or (hydroxyalkyl)amino side chain enhances anticancer activity of compounds [8][9][10][11]. There are also studies on novel derivatives with a 3,4-dihydronaphthalen moiety that exhibit anticancer activity [12][13]. Another important chemical group that is present in biologically active agents is hydrazone. It exhibits a broad spectrum of anticancer and antimicrobial activity [14][15][16][17][18][19]. The data obtained from the structure–activity relationships (SAR) analysis indicate that combined structures of pyrimidine, 3,4-dihydronaphthalene and alkylamine might have a synergistic anticancer effect.
As reported by the WHO, 18 million people are presently living with cancer; over 9 million people died from cancer in 2018. Due to the lack of effective and selectively acting anticancer therapies, these numbers are still increasing. The compounds based on the scaffold of pyrimidine exhibit a broad spectrum of pharmacological activity such as anti-inflammatory [1], antimicrobial [2,3], anti-HIV [2], antidiabetic [4] and anticancer activity [5,6,7]. The most recognized drugs based on analogs of pyrimidines are antibacterial (sulfadiazine, trimethoprim), antiviral (trifluridine, idoxuridine), anti-malarial (sulfadoxine), anti-HIV (Retrovir (zidovudine), stavudine), anti-tuberculosis (viomycin), anticancer (5-fluorouracil) agents. The data also show that the adjunction of a [(dialkylamino)alkyl]amino or (hydroxyalkyl)amino side chain enhances anticancer activity of compounds [8,9,10,11]. There are also studies on novel derivatives with a 3,4-dihydronaphthalen moiety that exhibit anticancer activity [12,13]. Another important chemical group that is present in biologically active agents is hydrazone. It exhibits a broad spectrum of anticancer and antimicrobial activity [14,15,16,17,18,19]. The data obtained from the structure–activity relationships (SAR) analysis indicate that combined structures of pyrimidine, 3,4-dihydronaphthalene and alkylamine might have a synergistic anticancer effect.
In the present paper, we analyzed and described the anticancer activity of newly designed compounds, a hybrid of pyrimidine–hydrazone moieties, dihydronaphthalene and alkylamine chain. All pyrimidine derivatives were evaluated in vitro for their antitumor activity against LoVo colon adenocarcinoma, LoVo/DX-resistant colon adenocarcinoma, MCF-7 breast cancer, A549 lung cancer, cervical cancer (HeLa), human leukemic lymphoblasts (CCRF-CEM) and human monocytic (THP-1) cell lines. We also evaluated their chemical and physical properties to determine the ability to be an orally active drug in humans. Basically, several different modes of action of the anticancer drug are recognized.
One of the possibilities is controlling transcription factors and polymerases where drugs interact directly with the protein bound to DNA. Another is the non-covalent binding of small molecules to double DNA structure by intercalation or interaction with the minor groove of nucleic acids. Based on the results of QSAR (Quantitative structure–activity relationship) studies, we propose the mode of action of designed compounds by binding to topoisomerase IIα (Topo IIα) in complex with DNA. Our study predicted their binding manner and the binding affinity towards Topo IIα by using molecular docking. It has been confirmed that Topo IIα is often overexpressed in different types of tumors, especially in the G2/M phase of the cell cycle and can be useful as a diagnostic marker [20][21]. Their inhibition leads to DNA double-strand breaks and apoptosis. To date, the FDA has approved etoposide, doxorubicin and mitoxantrone as Topo II-targeted chemotherapy agents [22].
One of the possibilities is controlling transcription factors and polymerases where drugs interact directly with the protein bound to DNA. Another is the non-covalent binding of small molecules to double DNA structure by intercalation or interaction with the minor groove of nucleic acids. Based on the results of QSAR (Quantitative structure–activity relationship) studies, we propose the mode of action of designed compounds by binding to topoisomerase IIα (Topo IIα) in complex with DNA. Our study predicted their binding manner and the binding affinity towards Topo IIα by using molecular docking. It has been confirmed that Topo IIα is often overexpressed in different types of tumors, especially in the G2/M phase of the cell cycle and can be useful as a diagnostic marker [20,21]. Their inhibition leads to DNA double-strand breaks and apoptosis. To date, the FDA has approved etoposide, doxorubicin and mitoxantrone as Topo II-targeted chemotherapy agents [22].
2. Chemistry
The starting point of synthesis was the reaction of 4,6-dichloro-pyrimidine (1) with hydrazine hydrate at room temperature. The resulting compound 4-chloro-6-hydrazinopyrimidine (2) was dissolved in ethanol and reacted with 6-methoxy-1-tetralone to obtain 4-chloro-6-[2-(6-methoxy-3,4-dihydronaphthalen-1(2H
)-ylidene)hydrazinyl]pyrimidine (3).
Compound 3 was refluxed in a nitrogen atmosphere at normal pressure withN
,N
-diethylethylenediamine orN
,N
-dimethylethylenediamine or 3-amino-1-propanol orN
,N
-dimethyl-1,3-propanediamine, respectively. The synthesis procedure is presented in (see the Methods section for details).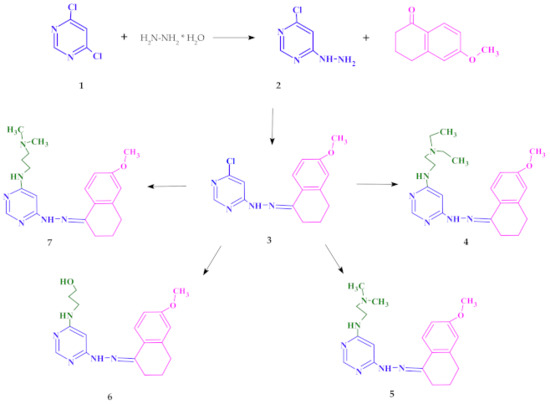
Scheme 1.
3. Lipophilicity and QSAR Study
The physicochemical properties like lipophilicity, aqueous solubility, and ADME (absorption, distribution, metabolism and excretion) properties [23] are important for drug candidate substances. While absorption, distribution, metabolism and excretion are related to pharmacokinetics, lipophilicity describes drug interactions with membranes. This is a very important aspect in understanding the mechanisms of transport of substances across the membrane into the cell, especially in the context of multidrug resistance (MDR). The ADME properties could be predicted by using theoretical computational techniques which various companies widely offer. In the presented paper, theoretical physicochemical properties of doxorubicin and all the tested compounds were determined based on the Lipinski and Veber rules on the SwissADME website [24] and were collected in . According to the Lipinski’s five rules, all the parameters determined for the tested compounds were in a good agreement with parameters for substances with potentially good pharmacokinetic properties (MW ≤ 500 Da, log P ≤ 5, number of hydrogen bond donors (NHD) ≤ 5 and number of hydrogen bond acceptors (NHA) ≤ 10).Table 1.
a
b
c
d
| Compound | Lipinski’s Rules | Veber’s Rules | |||||
|---|---|---|---|---|---|---|---|
| MW ≤ 500 | Log P ≤ 5 | NHD ≤ 5 | NHA ≤ 10 | Violations of Rules | NBR ≤ 10 | TPSA ≤ 140 | |
| Doxorubicin | 543.52 | 0.52 | 6 | 12 | 3 | 5 | 206.07 |
| 4 | 382.50 | 3.14 | 2 | 5 | 0 | 9 | 74.67 |
o/w
is an important factor for discovering and developing new effective therapeutics [25]. Among many methods of determining the log Po/w
coefficient, we chose the shake flask method, which is very simple and allows us to estimate the partition coefficient's relative values in the group of compounds under study. The values obtained for doxorubicin and the synthesized compounds 4, 5 and 7 are summarized in . The UV–Vis spectra are shown in (Supplementary Materials, could be found in https://www.mdpi.com/1422-0067/22/8/3825).
).Table 2.
o/w
o/w
o/w
| Compound | Log Po/w | Consensus log Po/w |
|---|---|---|
| Doxorubicin | –0.18 |
,34]. Etoposide binds to the binding site of topoisomerase and DNA with the free energy of binding equal to –44.6 kJ/mol. The aglycone part of the inhibitor localized between nucleic acid bases is involved in drug–DNA interactions (π–π interactions with thymine DT9 and guanine DG13). The podophyllotoxin moiety also contributes to drug–protein interactions and binds to the pocket created by Glu461, Gly462, Asp463, Arg487 and Gly488. The complex is also stabilized by van der Waals interactions and hydrogen bonds created with Gly462, Asp463 and guanine DG13 (see
and
and
).
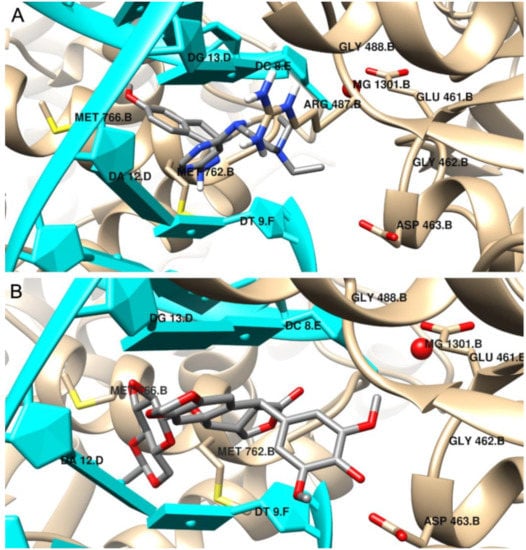
Figure 7.
A
B
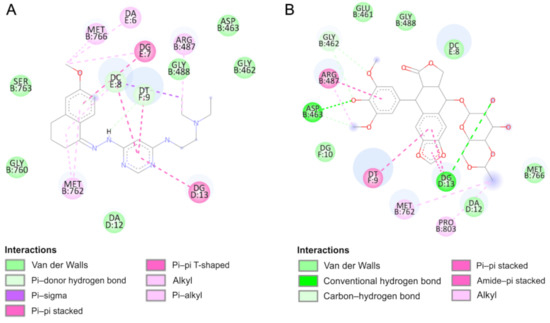
Figure 8.
A
B) etoposide.
Table 3.
| Compound | ΔGbinding (kJ/mol) |
Ki (µM) |
ΔGint (kJ/mol) |
ΔGvdw + ΔGhbond + ΔGdesolv (kJ/mol) |
ΔGel (kJ/mol) |
|||||
|---|---|---|---|---|---|---|---|---|---|---|
| 0.52 | ||||||||||
| 4 | −30.8 | 4.12 | −42.0 | −41.6 | −0.4 | |||||
| 4 | 1.05 | 3.14 | ||||||||
| 5 | 354.45 | 2.47 | 2 | 5 | 0 | 7 | 74.67 | |||
| 5 | 1.53 | 2.47 | 6 | 341.41 | 2.45 | 3 | 5 | 0 | 7 | 91.66 |
| 7 | 1.25 | 2.77 | 7 | 368.48 | 2.77 | 2 | 5 | 0 | 8 | 74.67 |
o/w
o/w
values. This may explain why the cytotoxic efficacy of the tested compounds compared to doxorubicin was higher. The best extraction into the organic layer was observed in the case of compound 4, which was followed by compounds 7 and 5 (). The 3D4D/QSAR model with a restricted docking protocol was also used to determine the biological activity of pyrimidine derivatives. We estimated the probability of their inhibitory activity towards topoisomerases I and II. We also analyzed anti-oxidant, DNA anti-metabolic and antimitotic activities. As presented in , compounds 4 and 7 exhibited very high probability of activity towards the Topo II enzyme. Compounds 4, 5 and 7 had antimitotic activity. QSAR analysis also indicated that compound 6 showed high probability of having anti-oxidant and DNA antimetabolic activity.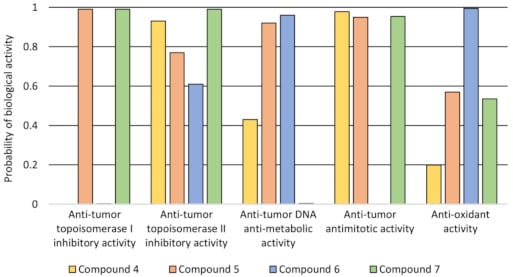
Figure 1.
4. Biological Evaluation
4.1. Cytotoxicity Assay
The high mortality rate among people with cancer is due to the lack of an effective anticancer drug that selectively targets cancer cells. In this article, we present new 4,6-substituted pyrimidine derivatives with anticancer activity. For a compound to exhibit pharmacological activity, its constituent parts should exhibit pharmacological activity. Properly selected structures contribute to finding a new group exhibiting activity and selectivity and low toxicity to healthy cells. The new structures are combinations of pyrimidine–hydrazone, dihydronaphthalene and alkylamine chain groups. Pyrimidine is a well-known aromatic, heterocyclic organic compound with many different biological functions [3][28][29].
Another functional group present in biologically active agents is hydrazone. It exhibits a broad spectrum of anticancer and antimicrobial activity [14][15][16][17][18][19]. On the other hand, there are reports in the scientific literature about dihydronaphthalene showing biological activity [30][31][32].
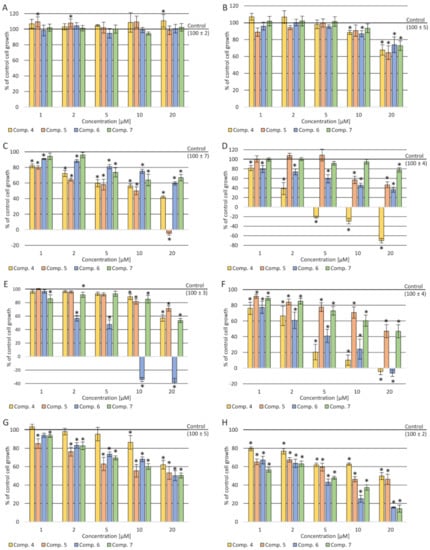
Figure 2.
A
B
C
D
E
F
G
H
p
4.2. Assessment of the Impact on the Transport Function of P-glycoprotein—Accumulation of Rhodamine 123 (Rod-123) in Cells
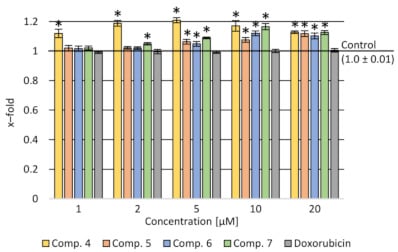
Figure 3.
p
4.3. Verification of Apoptotic and Necrotic Cell Death
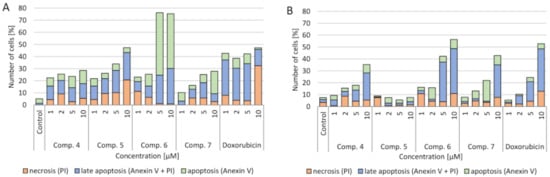
Figure 4.
A
B
4.4. Cell Cycle
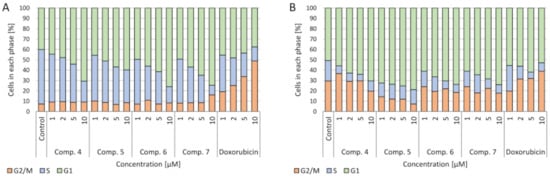
Figure 5.
A
B
4.5. Cell Migration
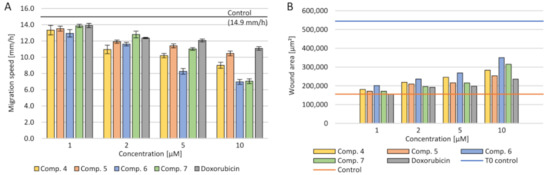
Figure 6.
A
B
5. Molecular Docking
We performed a molecular docking study to determine the binding mode of synthesized compounds to the Topo II/DNA complex (PDB ID 5GWK). We obtained the free energy of binding (ΔG
binding
) and the inhibition constants (K
i
) (see
Table 3). Additionally, the non-covalent interactions were characterized in detail. The docking protocol was validated by docking the co-crystalized ligand etoposide to the active site of Topo IIα. Our results are consistent with the earlier studies [33][34]. Etoposide binds to the binding site of topoisomerase and DNA with the free energy of binding equal to –44.6 kJ/mol. The aglycone part of the inhibitor localized between nucleic acid bases is involved in drug–DNA interactions (π–π interactions with thymine DT9 and guanine DG13). The podophyllotoxin moiety also contributes to drug–protein interactions and binds to the pocket created by Glu461, Gly462, Asp463, Arg487 and Gly488. The complex is also stabilized by van der Waals interactions and hydrogen bonds created with Gly462, Asp463 and guanine DG13 (see
). Additionally, the non-covalent interactions were characterized in detail. The docking protocol was validated by docking the co-crystalized ligand etoposide to the active site of Topo IIα. Our results are consistent with the earlier studies [33
| 5 | |||||
| −29.0 | 8.56 | −37.7 | −37.5 | −0.2 | |
| 6 | −26.3 | 24.36 | −36.3 | −35.5 | −0.8 |
| 7 | −27.0 | 15.77 | −37.0 | −36.8 | −0.2 |
| Etoposide | −44.6 | 0.02 | −50.9 | −50.6 | −0.3 |
According to the results, the most potent inhibitor of Topo IIα is compound 4 (inhibition constant, 4.12 µM). The pyrimidine ring is intercalated in double-stranded DNA and interacts via π–π- interactions with cytosine DC8, thymine DT9 and guanine DG13. The naphthalene moiety is involved in π-stacked interactions with guanine DG7 and π–alkyl interactions with Met762. Additionally, compound 4 forms van der Waals interactions with a number of polar, charged and hydrophobic amino acids of topoisomerase, namely, Gly462, Asp463, Gly488, Gly760 and Ser763.
As can be observed, the geometry of the designed ligands affects their binding properties and inhibitory activity. The data are presented in
and in the
. The
N
,
N
-diethylethylenediamine group with a
N
,
N
-dimethylethylenediamine fragment slightly changes the nature of interactions (compound 5). In this case, the naphthalene ring interacts via π–π stacking interactions with cytosine DC8, thymine DT9 and guanine DG13 and via π–σ interactions with adenine DA12. The van der Waals interactions of Glu461, Glu462, Gly760, Met762 and Tyr805 amino acid residues of Topo II and pyrimidine and
N
,
N
-dimethylethylenediamine fragments are present. The total intermolecular interaction energy for this complex is equal to −37.7 kJ/mol.
The results indicate that compound 6 is the weakest inhibitor of topoisomerase (ΔG
binding
= −26.3 kJ/mol), which is consistent with the quantitative structure–activity relationship study. It is probably related to the replacement of the branched chain of the 3-amino-1-propanol group, which interacts with the protein via hydrogen bonds with Asp463 and van der Waals interactions with Gly462, Ser464 and Gly488 amino acid residues. The naphthalene moiety exhibits a unique binding configuration, namely, interacts through π–σ interactions with guanine DG13, via a π–π stacked configuration with thymine DT9 and via π–alkyl interaction with adenine DA12. Its mechanisms of action can probably be different. QSAR studies showed that it has potential anti-oxidant and DNA antimetabolic activity. On the other hand, an in vitro study showed its strong proapoptotic properties.
Compound 7 can form hydrogen bonds with guanine molecules (DG10 and DG13). Additionally, the naphthalene ring is directly exposed to π–π interactions with thymine DT9 and guanine DG13. One alkyl interaction was observed between
N
,
N
-dimethyl-propanediamine chain and the Lys440 residue. In this case, three carbon–hydrogen bonds and van der Waals interactions were detected with Asp463, Pro485, Glu506 and Gly462 and Leu486, respectively.
