Ca2+-ATPases are key components of Ca2+ extrusion machinery and thus are pivotal for preservation of neuronal function.
- calmodulin
- calcium
- plasma membrane Ca2+-ATPase
1. Introduction
Ca
2+
-ATPases are key components of Ca
2+
extrusion machinery and thus are pivotal for preservation of neuronal function. Among three main calcium pumps, the plasma membrane Ca
2+
-ATPase (PMCA) and sarco/endoplasmic Ca
2+
-ATPase (SERCA) are known for decades while the secretory pathway Ca
2+-ATPase has been discovered in 2000s by two independent laboratories that described novel mutations leading to Hailey-Hailey disease [1][2][3]. All pumps have high affinity for Ca
-ATPase has been discovered in 2000s by two independent laboratories that described novel mutations leading to Hailey-Hailey disease [1,2,3]. All pumps have high affinity for Ca
2+
and function to restore cytosolic Ca
2+
concentration [Ca
2+
]
c
to the resting, nanomolar level following neuronal stimulations. They belong to the superfamily of mammalian P-type ATPases and are characterized by formation of a phosphorylated enzyme intermediate during catalytic cycle [1]. However, they have a low (~15%) degree of sequency identity [1], and differ in several other key features including tissue distribution, regulatory mechanisms, and contribution to neuronal Ca
2+ homeostasis. Each pump is encoded by multiple genes giving rise to a number of isoforms and further splice variants, which often possess distinguishable kinetic parameters and are dedicated to unique and highly regulated neural processes [4]. Naturally, the pumps share essential basic properties such as membrane topology, catalytic mechanism and probably the general features of 3D structure [5][6], although the structure of SPCA pump has not been solved yet (
homeostasis. Each pump is encoded by multiple genes giving rise to a number of isoforms and further splice variants, which often possess distinguishable kinetic parameters and are dedicated to unique and highly regulated neural processes [4]. Naturally, the pumps share essential basic properties such as membrane topology, catalytic mechanism and probably the general features of 3D structure [5,6], although the structure of SPCA pump has not been solved yet (
). The rapid expansion of the knowledge on pumps peculiar role, which run parallel to the advances in neuronal Ca
2+
signaling, led to the identification of several diseases associated either directly or indirectly with Ca
2+
pumps malfunction. Most of these defects have genetic background and the number of studies have been aimed to characterize their severity, effect on neuronal Ca
2+
homeostasis and signaling as well as neuronal survival. Besides known neuropathologies, defects in Ca
2+
pumps and alterations in the mechanisms regulating their activity may also produce subtle, tissue-specific disturbances that are not clinically manifested, yet they may affect neuronal machinery controlling and processing Ca
2+
signal.
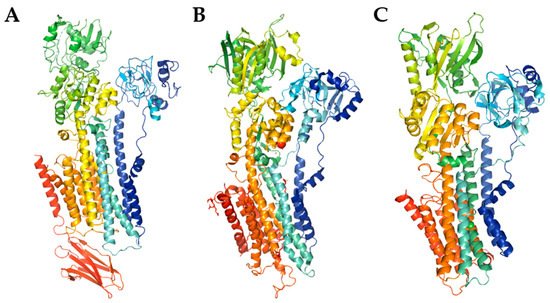
Figure 1.
The model of PMCA ((
A
), PDB entry code 6A69) and SERCA ((
B
), PDB entry code 3W5C) structures, and SPCA structure prediction using SWISS-MODEL (
C
). The cartoon models were generated with PyMOL.
2. Calmodulin—Ubiquitous Ca2+ Sensor in Neurons
Most commonly, detection and transduction of Ca
2+ signals in neurons are orchestrated by ubiquitous messenger called calmodulin. CaM is known as a relatively small (149aa; 16.7 kDa) and highly conserved calcium-binding sensor synthesized in all eukaryotic cells. It is particularity involved in synaptic signaling processes, neurotransmitter release and neuroplasticity by modulation (called “calmodulation”) of a large array of binding partners such as enzymes (e.g., adenylate cyclase, calcineurin, cyclic nucleotide phosphodiesterase, nitric oxide synthase, and certain kinases), transcription factors (e.g., CREB, NeuroD2, NFAT and MEF2) as well as various ion channels and transporters [7][8][9][10].
signals in neurons are orchestrated by ubiquitous messenger called calmodulin. CaM is known as a relatively small (149aa; 16.7 kDa) and highly conserved calcium-binding sensor synthesized in all eukaryotic cells. It is particularity involved in synaptic signaling processes, neurotransmitter release and neuroplasticity by modulation (called “calmodulation”) of a large array of binding partners such as enzymes (e.g., adenylate cyclase, calcineurin, cyclic nucleotide phosphodiesterase, nitric oxide synthase, and certain kinases), transcription factors (e.g., CREB, NeuroD2, NFAT and MEF2) as well as various ion channels and transporters [7,8,9,10].
In human, CaM is encoded by three independent genes CALM1, CALM2, CALM3 located on chromosomes 14q32.11; 2p21; and 19q13.32, respectively, which are collectively transcribed into at least eight mRNAs using different alternative polyadenylation signals (reviewed in [7][11]). Next, the resulting protein is susceptible to undergo various post-translational modifications, mainly phosphorylation on tyrosine (Thr26, Thr29, Thr44, Thr79, Tyr99, Thr117, and Tyr138) and serine (Ser81, and Ser101) sites [12]; acetylation of the N-terminal alanine [13]; trimethylation of the Lys115 [14]; and proteolytic cleavage at the C-terminal domain [15], all collectively regulating CaM biological activity. The crystal structure of mature CaM contains two independently folded lobes (N-lobe and C-lobe) connected by a flexible central α-helical linker, that differ by calcium affinity and kinetics of calcium dissociation. Each of these globular clusters can bind up to two free Ca
In human, CaM is encoded by three independent genes CALM1, CALM2, CALM3 located on chromosomes 14q32.11; 2p21; and 19q13.32, respectively, which are collectively transcribed into at least eight mRNAs using different alternative polyadenylation signals (reviewed in [7,11]). Next, the resulting protein is susceptible to undergo various post-translational modifications, mainly phosphorylation on tyrosine (Thr26, Thr29, Thr44, Thr79, Tyr99, Thr117, and Tyr138) and serine (Ser81, and Ser101) sites [12]; acetylation of the N-terminal alanine [13]; trimethylation of the Lys115 [14]; and proteolytic cleavage at the C-terminal domain [15], all collectively regulating CaM biological activity. The crystal structure of mature CaM contains two independently folded lobes (N-lobe and C-lobe) connected by a flexible central α-helical linker, that differ by calcium affinity and kinetics of calcium dissociation. Each of these globular clusters can bind up to two free Ca
2+
ions via a pair of helix-loop-helix motives (EF-hands) in a cooperative manner (K
d
= 5 · 10
−7
M to 5 · 10
−6 M) [16][17][18]. Because of subtle structural differences between these lobes resulting from evolutionary processes [19], EF hands in the C-lobe exhibit a three- to five times higher affinity for Ca
M) [16,17,18]. Because of subtle structural differences between these lobes resulting from evolutionary processes [19], EF hands in the C-lobe exhibit a three- to five times higher affinity for Ca
2+
. However, they possess slower rate of ion binding than the regions of EF hands located in the N-lobe, establishing the broad range of CaM sensitivity to the changes in calcium concentrations in the intracellular space [20]. CaM is susceptible to dramatic structural rearrangements via partially exposed hydrophobic patch on the C-terminal domain which may interact with CaM-binding proteins (CaMBPs) in a Ca
2+
-free (apo-CaM) state or in partially calcium-saturated forms (two Ca
2+ ions bound to the C-terminus) [16]. Up to date, over three hundred different calmodulin targets with specific binding sites and unique affinities for CaM, many of which located in the central nervous system (CNS) neurons [21], have been validated and extensively characterized [22]. The analysis of over 80 CaM complexes compiled in the Protein Data Bank (PDB) has revealed that CaM binding sites not always contain defined consensus sequence but rather share some common biochemical and biophysical properties such as high helix-forming propensities, positively charged binding region and the presence of hydrophobic anchor residues [8][22]. Thus, the classification of several CaM-binding motifs is determined by the spacing between these anchor residues as was extensively discussed by Mruk and colleagues [23]. As observed from sequence analysis of several CaMBPs, their IQ motif ([FILV]Qxxx[RK]Gxxx[RK]xx[FILVWY]) with highly conserved amino acid residues at positions 1, 2, 5, 6, 11, and 14 or IQ-like ([FILV]Qxxx[RK]Gxxxxxxxx) motif may also bind CaM in the presence or absence of Ca
ions bound to the C-terminus) [16]. Up to date, over three hundred different calmodulin targets with specific binding sites and unique affinities for CaM, many of which located in the central nervous system (CNS) neurons [21], have been validated and extensively characterized [22]. The analysis of over 80 CaM complexes compiled in the Protein Data Bank (PDB) has revealed that CaM binding sites not always contain defined consensus sequence but rather share some common biochemical and biophysical properties such as high helix-forming propensities, positively charged binding region and the presence of hydrophobic anchor residues [8,22]. Thus, the classification of several CaM-binding motifs is determined by the spacing between these anchor residues as was extensively discussed by Mruk and colleagues [23]. As observed from sequence analysis of several CaMBPs, their IQ motif ([FILV]Qxxx[RK]Gxxx[RK]xx[FILVWY]) with highly conserved amino acid residues at positions 1, 2, 5, 6, 11, and 14 or IQ-like ([FILV]Qxxx[RK]Gxxxxxxxx) motif may also bind CaM in the presence or absence of Ca
Considering the diversity of CaM interactions and its abundance in the brain (up to 100 μM range) [25], it seems rational to suspect that disruption of these multifunctional interactions regulating Ca
2+
-dependent intracellular signal transduction cascades may be implicated in the development of numerous neuropsychiatric disorders. Moreover, there is increasing evidence suggesting that pathophysiology of these states is intimately related to the disturbed neuronal calcium homeostasis also mediated by ATP-driven pumps located in the plasma membrane, in the membranes of the endoplasmic reticulum (ER), or Golgi compartments.
3. Plasma Membrane Ca
2+
-ATPase (PMCA)—The Only Calcium Pump Directly Regulated by Calmodulin
PMCA is one of the most important and sensitive players in maintaining of low resting Ca
2+
concentration, and ensuring a fast recovery of [Ca
2+
]
c to the basal level following neuronal excitation [26]. The enzyme was first described by Schatzmann in 1960s as ATP-powered mechanism that removes calcium from red blood cells [27], whereas further studies revealed the presence of PMCA in other cells, including neurons [28][29][30] Structurally, PMCA comprises of ten transmembrane segments with N- and C- terminal tails both located on the cytosolic site [31]. Most of the regulatory regions including acidic phospholipids, protein kinase C (PKC), protein kinase A (PKA) and the crucial natural activator—CaM, are located at the C- terminus. The important regulatory role of CaM in stimulating of PMCA is associated with increasing the affinity of the pump for calcium and the maximum rate of calcium extrusion. In the activation process, CaM removes the auto-inhibitory C-terminal domain from the active site and releases the enzyme from auto-inhibition [32]. It is also worth mentioning that PMCA is so far the only known calcium pump directly activated by CaM [26].
to the basal level following neuronal excitation [26]. The enzyme was first described by Schatzmann in 1960s as ATP-powered mechanism that removes calcium from red blood cells [27], whereas further studies revealed the presence of PMCA in other cells, including neurons [28,29,30] Structurally, PMCA comprises of ten transmembrane segments with N- and C- terminal tails both located on the cytosolic site [31]. Most of the regulatory regions including acidic phospholipids, protein kinase C (PKC), protein kinase A (PKA) and the crucial natural activator—CaM, are located at the C- terminus. The important regulatory role of CaM in stimulating of PMCA is associated with increasing the affinity of the pump for calcium and the maximum rate of calcium extrusion. In the activation process, CaM removes the auto-inhibitory C-terminal domain from the active site and releases the enzyme from auto-inhibition [32]. It is also worth mentioning that PMCA is so far the only known calcium pump directly activated by CaM [26].
In mammals, four isoforms of PMCA (PMCA1-PMCA4), structurally similar to each other, have been found [4] but their expression depends on cell type (
). The PMCA1 and PMCA4 are widely expressed in virtually all animal tissues and both play a house-keeping role.
Table 1.
Properties of PMCA isoforms. Modified based on [1].
| PMCA1 | PMCA2 | PMCA3 | PMCA4 | |
|---|---|---|---|---|
| Tissue Distribution | Ubiquitous | Restricted (brain) |
Restricted (brain) |
Ubiquitous |
| Developmental Expression/Switch | Isoform switch fetal/adult | Isoform switch fetal/adult | Isoform switch fetal/adult | Isoform switch fetal/adult |
| Affinity CaM (Kd nM) | 40–50 | 2–4 | 8 | 3–40 |
Expression of PMCA2 and PMCA3 is highly restricted to excitable cells and their high concentration has been detected in the CNS [4]. PMCA2 is especially abundant in cerebellar Purkinje cells and granule cells, but it also localizes to the cerebral cortex and hippocampus [33]. PMCA3, in turn, is present predominantly in cerebellar granule cells and in the choroid plexus [34] what suggests its role in generation and release of cerebrospinal fluid. Additionally, PMCA isoforms are characterized by distinct calmodulin sensitivity (
) and specific kinetic properties. PMCA2 and PMCA3 are referred to as “fast” isoforms due to their high basal activity and high affinity for CaM, whereas PMCA1 and PMCA4 are much slower despite their strong stimulation by CaM [1]. It has been suggested that the cell response to a physiological stimulus depends on significant differences in the kinetic parameters of the individual isoforms. In the brain, distribution of PMCA isoforms clearly alters during development, what may indicate their specific role in embryogenesis and further in postnatal period [35].
