The UK can be proud of the fact that numerous native breeds of sheep have been developed here that possess unique phenotypic features and excellent productivity and are utilized throughout the world. Their remarkable popularity and further sustainable breeding on grass pastures of British Isles and elsewhere can benefit from genomic applications. At present, there is a rich arsenal of genetic and genomic resources, tools and applications used for livestock assessment, breeding and production including, first of all, genetic profiling of diverse breeds, and search for quantitative trait loci (QTLs) and candidate genes in farm animals. These genomic advances facilitate breed improvement and understanding of the genetic processes in the course of domestication and breed evolution.
- British sheep breeds
- diversity
- genetics
- genomics
- conservation
- adaptation
1. Introduction
1. Introduction
Sheep farming has been an important sector of the UK’s economy and rural life for many centuries. It is the favored source of wool, meat and milk products. In the era of exponential progress in genomic technologies, we can now address the questions of what is special about UK sheep breed genotypes and how they differ genetically form one another and from other countries. We can reflect how their natural history has been determined at the level of their genetic code and what traces have been left in their genomes because of selection for phenotypic traits. These include adaptability to certain environmental conditions and management, as well as resistance to disease. Application of these advancements in genetics and genomics to study sheep breeds of British domestic selection has begun and will continue in order to facilitate conservation solutions and production improvement.Sheep farming has been an important sector of the UK’s economy and rural life for many centuries. It is the favored source of wool, meat and milk products. Their ubiquity in the scientific literature dates back at least to William Youatt
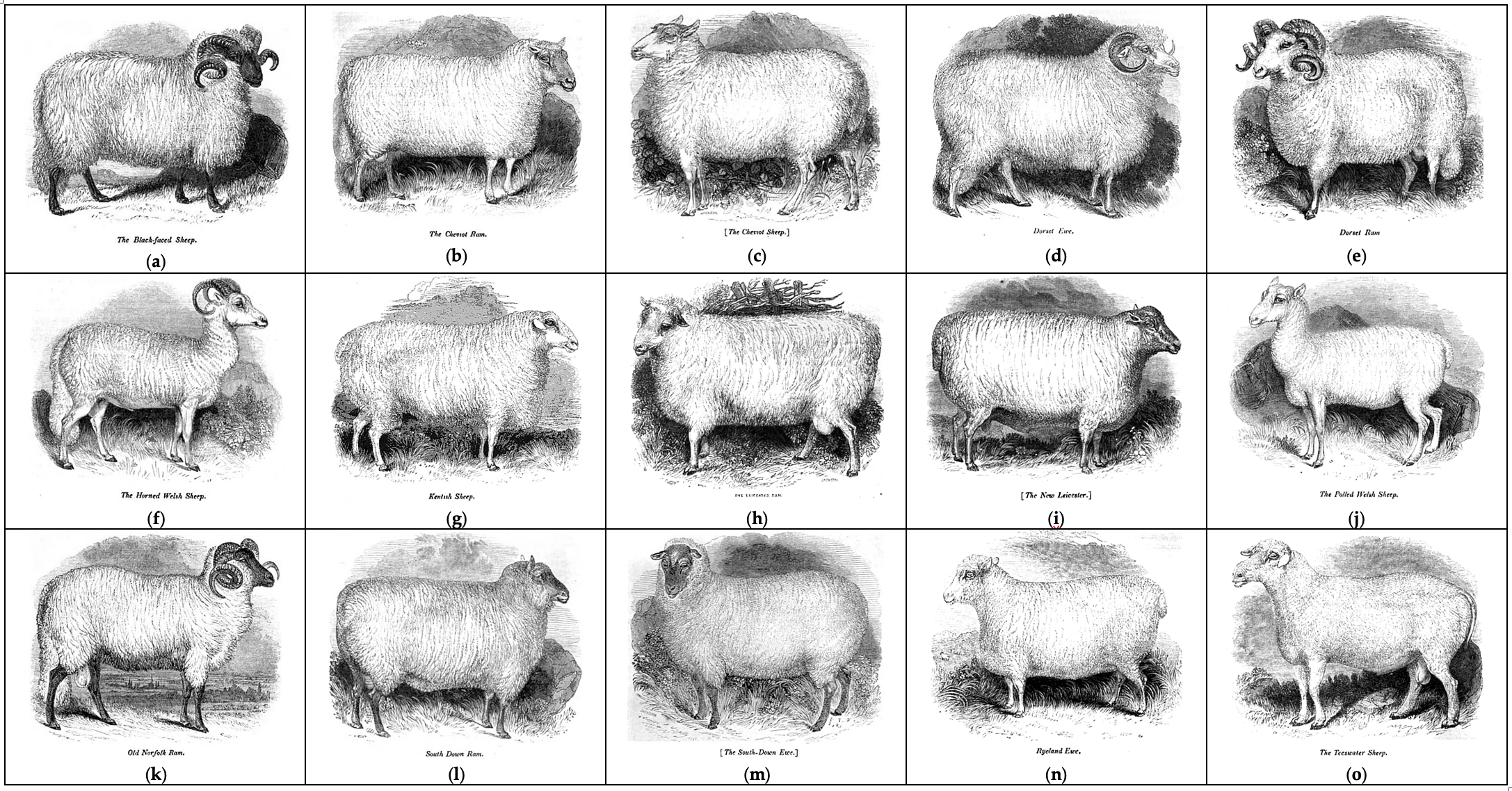
(Figure 1) who provided an impressive overview of sheep breeding, management, veterinary aspects and their relevance to everyday life.
2. Genetic Diversity, QTL and Candidate Gene Characterization
Figure 1. Old British sheep breeds as shown in the 1837 treatise by William Youatt
[1]: (a) Blackfaced sheep, (b) Cheviot ram, (c) Cheviot sheep, (d) Dorset ewe, (e) Dorset ram, (f) Horned Welsh sheep, (g) Kentish sheep, (h) Leicester ram, (i) New Leicester, (j) Polled Welsh sheep, (k) Old Norfolk ram, (l) South Down ram, (m) South Down ewe, (n) Ryland ewe, and (o) Teeswater sheep.
In the era of exponential progress in genomic technologies, we can now address the questions of what is special about UK sheep breed genotypes and how they differ genetically form one another and from other countries. We can reflect how their natural history has been determined at the level of their genetic code and what traces have been left in their genomes because of selection for phenotypic traits. These include adaptability to certain environmental conditions and management, as well as resistance to disease. Application of these advancements in genetics and genomics to study sheep breeds of British domestic selection has begun and will continue in order to facilitate conservation solutions and production improvement.
It requires a major undertaking to evaluate genetically most widespread industrial breeds [2], such as the Texel in sheep [3]. However, more and more attention is being drawn to surveying and analyzing local livestock breeds. This is due to their adaptive properties, as reflected in their genomic structure, and their potential to improve performance, resistance and environment impact of commercial herds (e.g., [2][4][5]).
2. Geographical Distribution
2.1. British Isles
Sheep breed gene pool of British Isles can be subdivided into geographical groups (Figure 2) according to their origin from England (Border Leicester; Clun Forest; Dorset Horn, Figure 1d,e; English Leicester, Figure 1h,i; Romney; Southdown, Figure 1l,m; Suffolk; Wiltshire Horn; etc.), Isle of Man (Manx Loaghtan), Scotland (Cheviot, Figure 1b,c; Scottish Blackface, Figure 1a; etc.), Wales (Badger Face Welsh Mountain, Balwen Welsh Mountain, Beulah Speckled Face, Black Welsh Mountain, Brecknock Hill Cheviot, Hill Radnor, Improved Welsh Mountain, Improved Welsh Mountain, Kerry Hill, Llandovery Whiteface, Llanwenog, Lleyn, Talybont Welsh, Welsh Hardy Speckled Faced, Welsh Mountain-Hill Flock, etc.), and Ireland (Galway).
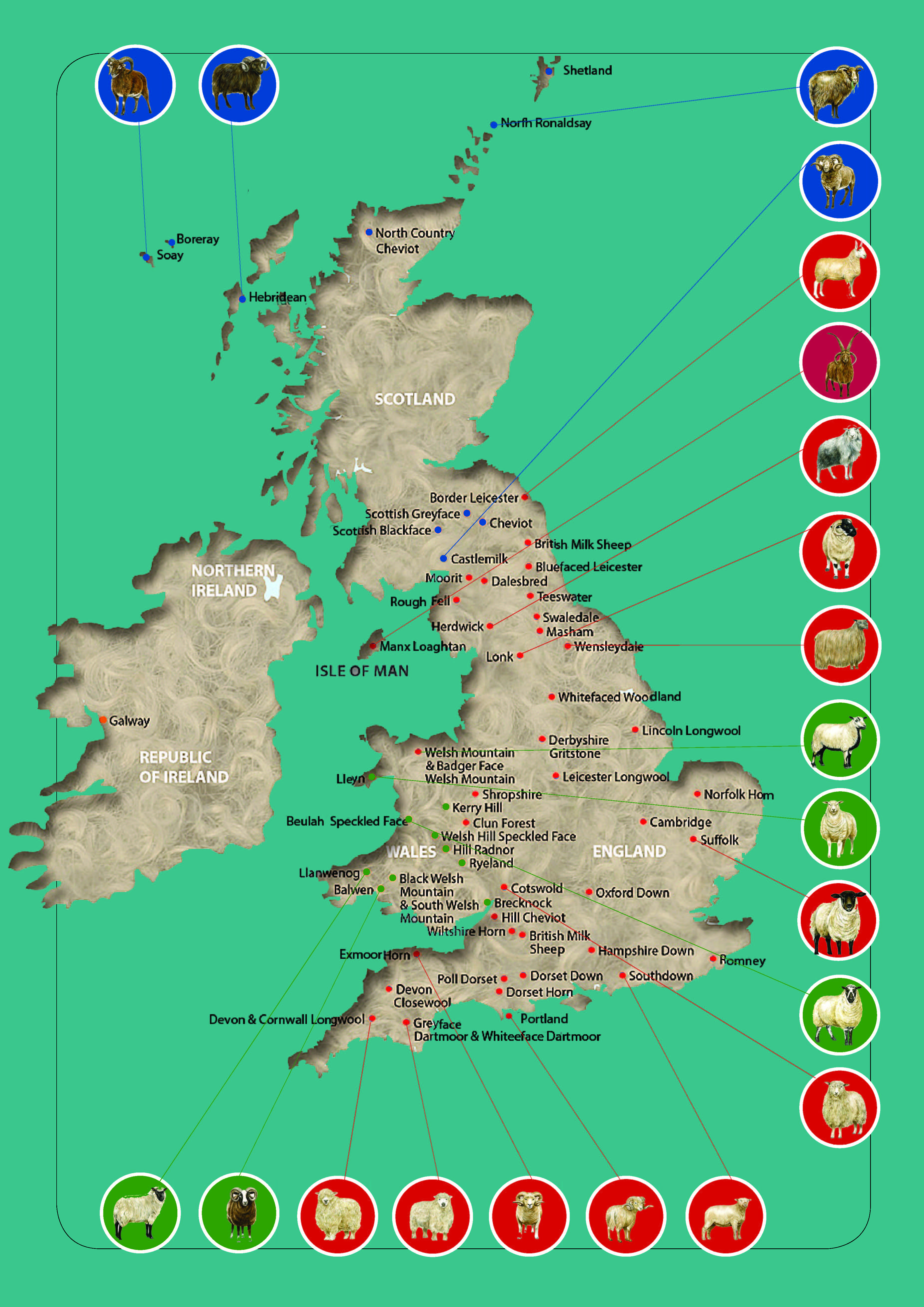
Figure 2. Geographical distribution of native sheep breeds in the British Isles. Modified from
The geographical concentration (endemism) of the British sheep breeds was suggested to be a major risk factor for breed endangerment
.
2.2. Outside Britain
British sheep breeds are common in other parts of the world and have been exported from the UK to other countries to create new breeds and improve the extant ones, with Australian, USA and European breeds being prominent examples [9]
[6][5][10]. As in other examples, imports into the former USSR can be mentioned when British breeds served as a basis for developing new breeds, e.g., Gorky and Russian Long-wool
[11]
[12]
[13]
[14]
[15]
3. Genomic Applications
[16]. In particular, British meat sheep breeds have had a significant impact on the development of native gene pool breeds for semi-fine sheep production in Russia and former USSR countries in the last century [17]
[18]
[19]
[20]
[21] (see Figure 3).

Figure 3. Examples of Soviet sheep breeds produced using British breeds and presented at the All-Union Agricultural Exhibition (AUAE): (a) Mikhnov × Lincoln crossbred ewe (year of birth, 1938; wool fineness, 44–46; body weight, 104 kg; AUAE breed champion, 1940)
[1122]; (b) Kuibyshev ram (year of birth, 1950; body weight, 152 kg; wool yield, 6.3 kg; wool length, 21.0 cm; wool fineness, 46; AUAE champion, 1954)
[1223]; (c) Pechora ram (year of birth, 1951; body weight, 94 kg; wool yield, 5.62 kg; wool length, 12.0 cm; wool fineness, 50; 1st degree AUAE certificate, 1954)
[1216]; (d) Russian Long-wool ram (year of birth, 1951; body weight, 126 kg; wool yield, 9.5 kg; wool length, 24.0 cm; wool fineness, 44; AUAE champion, 1954)
[1224]; (e) Lithuanian Black-headed ram (year of birth, 1949; body weight, 131 kg; wool yield, 6.2 kg; wool length, 11.0 cm; wool fineness, 56–50; 1st degree AUAE certificate, 1954)
[1224]; (f) Gorky ewe (year of birth, 1951; body weight, 79 kg; wool yield, 5.3 kg; wool length, 10.0 cm; wool fineness, 56; 2nd degree AUAE certificate, 1954)
[1222]; (g) Latvian Dark-headed ewe (year of birth, 1949; body weight, 102 kg; wool yield, 6 kg; wool length, 10.5 cm; wool fineness, 56–50; AUAE breed champion, 1954)
[1215]; (h) Estonian Black-headed ram (year of birth, 1952; body weight, 85 kg; wool yield, 4.8 kg; wool length, 11.0 cm; wool fineness, 56–50; 2nd degree AUAE certificate, 1954)
[12].
3. Phenotypic Diversity
Many of British breeds are unique in terms of phenotypic traits and adaptation to local conditions [22] but nonetheless may contribute to the commercial herds. For instance, Bluefaced Leicester is believed to be most distinguished breed in the UK as it sires almost half of the commercial hybrid progeny
In terms of peculiar adaptability, specific phenotypes and features of the British sheep breeds, some of them are lowland breeds (Suffolk, Wiltshire Horn), some are hill breeds (Cheviot, Scottish Blackface, Brecknock Hill Cheviot, Hill Radnor, Welsh Mountain-Hill Flock), and some are upland breeds (Beulah Speckled Face, Llandovery Whiteface, Llanwenog), while some others are adapted to variable conditions (Clun Forest, Badger Face Welsh Mountain, Kerry Hill, Lleyn)
[23]. An extraordinary example of adaptation is the North Ronaldsay breed localized on an Orkney island, whose copper-deficient and high-sodium diet is based predominantly on the seaweed Laminaria [24].
Body size varies from small (Black Welsh Mountain) to medium (Clun Forest, Llanwenog, Lleyn, Welsh Mountain-Hill Flock) to large (Border Leicester, Romney, Wiltshire Horn, Talybont Welsh). Some breeds are horned (Dorset Horn, Wiltshire Horn, Scottish Blackface), others are hornless (Border Leicester, Romney, Beulah Speckled Face, Kerry Hill, Lleyn), and in some others only males have horns (Cheviot, Black Welsh Mountain, Balwen Welsh Mountain, Brecknock Hill Cheviot, Llandovery Whiteface, Welsh Mountain-Hill Flock). The Border Leicester breed has a characteristic “Roman nose”.
Most breeds are used for producing lamb meat, while some (Clun Forest) are dual purpose and can be used for meat, wool and milk. There are long-woolled breeds (e.g., Romney whose fleece is suitable for carpets), and others (e.g., Wiltshire Horn) naturally shed wool, not requiring shearing. Females of some breeds (Dorset Horn, Hill Radnor) have a slightly shorter gestation period allowing 1.5 lambs a year. Some breeds (Romney, Balwen Welsh Mountain, Lleyn) have a low disease risk. Previously, British sheep breeds and flocks were assessed, on the base of prion protein (PrP) genotypes, for risk of scrapie, also known as prion disease and transmissible spongiform encephalopathy [25]
that had implications for breeding programs. As one more example, the Dorset breed was found to be a carrier of a single polymorphism mutation at the callipyge (CLPG) locus causing the muscle hypertrophy phenotype
[30].
4. Genetic Diversity, QTLs and Candidate Gene Characterization
To characterize genetic structure and diversity in the sheep, various molecular markers were previously utilized, including microsatellites (e.g.,
3.1. SNPs
[24][35]; see for review [2216][3617]), mtDNA (e.g., [36][37][38]) and endogenous retroviruses
[396]. For example, in a study of three English breeds [225]
[3524], it was shown that they were clearly distinguished relative to one another for ten microsatellite loci. One breed, the Herdwick, was unique for high frequency of the R0 retrotype indicative of a primitive genome that is absent in the mainland UK breeds and known only for few other non-British breeds.
Using microsatellite markers, QTLs associated with muscle depth were characterized in British commercial terminal sire sheep including the Suffolk breed
[40]. One QTL for muscle depth was verified in Suffolk sheep on chromosome 1.
Since ewe prolificacy was associated with certain mutations in the BMP15 and GDF9 candidate genes, it was explored in UK and Ireland sheep by their genotyping for these alleles
. Three mutations had large effects on ovulation rate in the Cambridge and Belclare (of Irish origin) breeds, with two alleles being transferred from the Lleyn breed (of Welsh origin) and one from a High Fertility line in Ireland.
Genetic resistance to nematode infection is an important target of selective breeding for this trait in the UK. This was studied within a purebred Scottish Blackface flock by partial resequencing genes in the Major Histocompatibility Complex (MHC) class II region [42]. Causal mutant alleles at the DRB1 and DQB2 loci were identified that were associated with this trait. Single nucleotide polymorphisms (SNPs) in three other candidate genes for nematode resistance and body weight were examined in populations of domestic Scottish Blackface and free-living Soay ewe lambs, and a nominally significant association between an IL23R SNP and body weight was found [43].
Other examples of candidate genes, for example, associated with ewe mature weight are TMEM8B and SPAG8 that showed picks of a signature of selection at single SNPs in four sheep breeds, the Suffolk among them [44].
5. Genomic Applications
With the advent of next generation sequencing (NGS) technologies, SNP panels and a whole genome sequence draft became available for the sheep by 2010 [45]
[46] that can also be used for querying genomic features of British breeds. The remarkable milestone in this field was the annotated sheep genome sequence Oar v3.1 published in 2014
[47]. Another improved assembly, Oar_v4.0, was produced using PBJelly 2 software
[48]. The latest genome assemblies were generated in 2017 and 2020, and designated Oar_rambouillet_v1.0 (sheep reference genome;
[49]) and ASM1117029v1
[50], respectively.
These state-of-the-art resources are crucial for genetic improvement of the existing sheep flock by implementing genome-wide association studies (GWAS; e.g.,
3.2. Whole Genome Sequencing
[323]), analysis of quantitative traits and genomic selection [5125]. However, a key prerequisite for these applications is a thorough examination of genetic structure and variation within and between sheep breeds including the British ones. This information also helps elucidate domestication pathways, breed formation and population history [45]. In particular, insight into demographic history of breeds can provide a set of genetic markers for obtaining individual genomic estimated breeding values (GEBV) (i.e., genomic selection) and their applicability to other populations [4]. Efficacy of genomic and marker-assisted selection, and QTL spotting via GWAS depends on knowledge of population structure and origin [4].
After marker validation, genetic or genomic selection is feasible when targeting, for example, such sheep traits as footrot resistance [3] and mature body weight [44]. For genomic selection implementation, a genotyped reference population is built for GEBV evaluation. As low heritability and polygenic nature is inherent in selected quantitative traits, genomic selection hopefully improves selection response if compared to conventional best linear unbiased prediction-assisted selection [3].
There are two major collaborative sheep genomics groups, the International Sheep Genomics Consortium [46] and an Australia- and New Zeeland-based project, SheepGenomesDB [52]. Another beneficiary group is the Ovine Functional Annotations of Animal Genomes (FAANG) Project [53][54][55]. Studies within the framework of the FAANG [55][56] and related projects [57] also produced sheep genome datasets including those for British breeds.
5.1. SNPs
Use of multiple SNP markers has substantially enhanced analysis of genetic diversity and population history [22][37][58][59][60][61], especially thanks to the sheep HapMap project [4][45][46][62][63][64][65]. For instance, in a genome-wide survey of SNP variation [45], it was demonstrated that the British Suffolk genetically differentiated from two American Suffolk subpopulations, whereas the genetic structure of Australian Poll Dorset and American Dorsets was also different. In another research of genetic structure and admixture in terminal sire breeds in the USA using Applied Biosystems Axiom Ovine Genotyping Array (50K) and Illumina Ovine SNP50 BeadChip, the Suffolk, Hampshire, Shropshire and Oxford (terminal) sheep were genotyped along with the Rambouillet (or the French Merino; dual purpose) sheep [66]. There was a clear-cut divergence between the Suffolk sheep from two different US regions. The Hampshire, Suffolk, and Shropshire breeds demonstrated the greatest admixture. Relative to sheep from other world regions, the US terminal breeds of British origin formed a separate cluster suggesting their genetic distinctiveness.
The earliest research of SNP-based diversity in UK sheep showed genetic distinctiveness of three English native hill breeds examined at three SNP loci associated with phenotypes [22][35]. In a broader study of 18 Welsh local breeds as a selected segment of the UK’s sheep germplasm [4], the Illumina OvineSNP50 array was employed to examine genetic structure of these breeds. A similar methodology was exploited to elucidate genetic diversity and genome selection in the Suffolk, Rambouillet and three Rambouillet-related breeds from the USA [67]. The Suffolk sheep were clearly distinguished from the four others in terms of diversity and differentially selected genome regions.
SNPs have also become genetic markers of choice in searching for QTLs and conducting GWAS in sheep (e.g., [3][68][69][70][71][72]). The Illumina OvineSNP50 chip was utilized for a GWAS and regional heritability mapping (RHM) to identify QTLs for nematode resistance and body weight in Scottish Blackface lambs [73]. Strong associations were found on chromosomes 14 and 6 for nematode resistance, and on chromosome 6 for body weight. An additional RHM study in three European populations (including Scottish Blackface) revealed other QTLs for nematode resistance, with one on chromosome 20 being the most significant and located close to MHC, as a functional candidate for this trait [74]. In the follow-up investigation [75], accuracy of genomic prediction within and across populations for nematode resistance and body weight was assessed in two British purebred (Scottish Blackface, British Texel) and two non-British backcross populations. GEBV were definitely better within populations that points out a more accurate genomic prediction in closely related sheep than across breeds. Later, using a 932-SNP assay, an independent validation search for nematode resistance QTLs in three sheep breeds (including Scottish Blackface and Suffolk) suggested that inconsistency of SNP effects may occur in different populations [76].
The same Illumina OvineSNP50 genotype panel was an effective tool for investigating runs of homozygosity (ROHs) and selection signatures in six commercial European meat breeds including the Suffolk sheep (of Irish population) [77]. The Suffolk breed showed a distinct population structure different from five other breeds. Moreover, the Suffolk sheep were the least admixed, although they formed two non-overlapping clusters, one of them being a subpopulation of New Zealand origin. The Irish Suffolk population was more abundant in ROHs, suggesting its smaller effective population size both in recent and past generations, and a higher relatedness among this breed. The Suffolk (along with the Beltex) had the largest number of putative selective sweeps.
5.2. Whole Genome Sequencing
Further development of NGS platforms and reduction of their cost make it possible to implement whole genome sequencing for numbers of individuals within one or more species. Whole genome sequences seem to provide ultimate evaluation of genetic variability and candidate mutations that can be further used for GWAS, sequence genotype imputation and genomic prediction improvement as a component of genomic selection
[51]
Using whole genome sequences of 21 Chinese native sheep breeds, Yang et al.
[7832] identified candidate genes, pathways and gene ontology categories presumably related to high-altitude and arid environments.
Naval-Sanchez et al. [57] sequenced 43 worldwide sheep breeds and functionally annotated their whole genome sequences, demonstrating that selection sweeps correspond to coding genes, proximal regulatory elements and active transcription sites, and suggesting that remodeled gene expression could play an important evolutionary role in sheep breed diversification.
On the basis of whole genome sequences for 145 wild and domestic sheep and goat samples, Alberto et al.
[5938] found selective sweeps that led to domestic breed divergence as well as genomic signatures for convergent domestication in two related species.
Using SNP and whole genome sequence data for a large worldwide sample collection of wild and domestic animals, Chen et al. [65] found that in sheep there might be an accelerated genetic drift vs. reduced directional selection on X chromosome as compared to autosomes.
The ongoing Australia- and New Zeeland-based project SheepGenomesDB is aimed at sequencing 453 (Run1) and 935 (Run2) animals, which is supposed to cover a global sheep breed diversity in order to identify causative mutations and facilitate genomic selection
6. British Sheep Genome Studies
4. British Sheep Genome Studies
To date, there has not been launched an overall, comprehensive genetic/genomic survey of the British sheep gene pool. Certain British native breeds have been part of either diversity studies using different molecular markers or whole genome sequences and often a limited number of samples per breed. For example, using the Illumina HiSeq 2000 platform, three individual animals, each representing one Welsh sheep breed (Hardy Speckled Faced, Dollgellau and Tregaron Welsh Mountain), were sequenced in order to explore their demographic history
Recently, more whole genome sequences were generated for separate British breeds. For instance, the Illumina whole genome sequences for seven Cambridge sheep and one British du Cher individual were obtained in a comparative study including also the Romanov and two Iranian breeds
[7952]. The Cambridge breed was genetically different, while the British du Cher being close to the Romanov breed. A higher number of short ROHs was detected in the Cambridge sheep and a lower number of long ROHs in the British du Cher, meaning a lower recent inbreeding in the latter breed.
Whole genome sequences of 17 Poll Dorset sheep was compared to those from two Tibetan breeds
[8053]. Selection signatures were identified that include candidate genes putatively associated with hypoxia responses, meat traits and disease resistance.
There are also international projects that have generated whole genome sequences for British and non-British breeds and can be served as data sources for further studies
[5629][7750][8154][8255]. In a recent global genomic survey of wild sheep and domestic breeds
PDGFD
XYLB
TSHR
SGCZ
PDGFD
5. Conclusions
7. Conclusions
The UK sheep landscape is characterized by abundant breed history and genetic diversity, suggesting its preservation for sustainable use and advanced research. The latter can rely upon state-of-the-art molecular and genomic tools including SNPs and whole genome sequencing. This opens further opportunities to elucidate breed ancestry, variability, and genetic merit of particular markers and candidate genes for adaptation, genomic selection and production improvement. Further research is anticipated to understand comprehensively how genomic landscape of the British sheep has contributed to the fact that over the centuries they have thrived to current numbers and value, delighting our eyes on the green landscapes of today’s Great Britain.
References
- Youatt, W. Sheep: Their Breeds, Management, and Diseases: To Which Is Added the Mountain Shepherd’s Manual; Baldwin and Cradock: London, UK, 1837.Zamorano, M.J.; Ruiter, J.; Townsend, S.; Cruickshank, R.; Bruford, M.; Byrne, K.; Rodero, A.; Vega-Pla, J.L. Polimorfismos de DNA en las razas ovinas Merino y Churra lebrijana: DNA polymorphisms in Merino and Churra lebrijana Sheep breeds. Arch. Zoot. 1998, 47, 267–272.
- Yurchenko, A.; Yudin, N.; Aitnazarov, R.; Plyusnina, A.; Brukhin, V.; Soloshenko, V.; Lhasaranov, B.; Popov, R.; Paronyan, I.A.; Plemyashov, K.V.; et al. Genome-wide genotyping uncovers genetic profiles and history of the Russian cattle breeds. Heredity 2018, 120, 125–137.Peter, C.; Bruford, M.; Perez, T.; Dalamitra, S.; Hewitt, G.; Erhardt, G.; Econogene Consortium. Genetic diversity and subdivision of 57 European and Middle-Eastern sheep breeds. Anim. Genet. 2007, 38, 37–44.
- Mucha, S.; Bunger, L.; Conington, J. Genome-wide association study of footrot in Texel sheep. Genet. Sel. Evol. 2015, 47, 35.Lawson Handley, L.J.; Byrne, K.; Santucci, F.; Townsend, S.; Taylor, M.; Bruford, M.W.; Hewitt, G.M. Genetic structure of European sheep breeds. Heredity 2007, 99, 620–631.
- Beynon, S.E.; Slavov, G.T.; Farré, M.; Sunduimijid, B.; Waddams, K.; Davies, B.; Haresign, W.; Kijas, J.; MacLeod, I.M.; Newbold, C.J.; et al. Population structure and history of the Welsh sheep breeds determined by whole genome genotyping. BMC Genet. 2015, 16, 65.Russo-Almeida, P.A. Diversidade Genética e Diferenciação das Raças Portuguesas de Ovinos com Base em Marcadores de DNA–Microssatélites: Uma Perspectiva de Conservação. Tese de Doutorado em Ciência Animal, Universidade de Trás-os-Montes e Alto Douro, Vila Real, Portugal, 2007.
- Weigend, S.; Romanov, M.N. The World Watch List for Domestic Animal Diversity in the context of conservation and utilisation of poultry biodiversity. Worlds Poult. Sci. J. 2002, 58, 519–538.Bowles, D.; Carson, A.; Isaac, P. Genetic distinctiveness of the Herdwick sheep breed and two other locally adapted hill breeds of the UK. PLoS ONE 2014, 9, e87823.
- Green, K. Shaggy sheep stories. Ctry. Life 2017, 121 (13), 68–72.Bowles, D. Recent advances in understanding the genetic resources of sheep breeds locally-adapted to the UK uplands: Opportunities they offer for sustainable productivity. Front. Genet. 2015, 6, 24.
- The Natural Fibre Company. Meet the Animals. Available online: https://www.thenaturalfibre.co.uk/meet-the-animals (accessed on 3 February 2021).Bruford, M.; Townsend, S.J. Mitochondrial DNA diversity in modern sheep: Implications for domestication. In Documenting Domestication: New Genetic and Archaeological Paradigms; Zeder, M.A., Bradley, D.G., Emshwiller, E., Smith, B.D., Eds.; University of California Press: Berkeley, CA, USA, 2006; pp. 307–317.
- Carson, A.; Elliott, M.; Groom, J.; Winter, A.; Bowles, D. Geographical isolation of native sheep breeds in the UK-Evidence of endemism as a risk factor to genetic resources. Livest. Sci. 2009, 123, 288–299.Pariset, L.; Mariotti, M.; Gargani, M.; Joost, S.; Negrini, R.; Perez, T.; Bruford, M.; Ajmone Marsan, P.; Valentini, A. Genetic diversity of sheep breeds from Albania, Greece, and Italy assessed by mitochondrial DNA and nuclear polymorphisms (SNPs). Sci. World J. 2011, 11, 1641–1659.
- World Watch List for Domestic Animal Diversity, 3rd ed.; Scherf, B.D. (Ed.) FAO: Rome, Italy, 2000.Lv, F.H.; Peng, W.F.; Yang, J.; Zhao, Y.X.; Li, W.R.; Liu, M.J.; Ma, Y.H.; Zhao, Q.J.; Yang, G.L.; Wang, F.; et al. Mitogenomic meta-analysis identifies two phases of migration in the history of Eastern Eurasian sheep. Mol. Biol. Evol. 2015, 32, 2515–2533.
- FAO. Domestic Animal Diversity Information System (DAD-IS). Available online: http://www.fao.org/dad-is/en/ (accessed on 14 February 2021).Chessa, B.; Pereira, F.; Arnaud, F.; Amorim, A.; Goyache, F.; Mainland, I.; Kao, R.R.; Pemberton, J.M.; Beraldi, D.; Stear, M.J.; et al. Revealing the history of sheep domestication using retrovirus integrations. Science 2009, 324, 532–536.
- Deikhman, E.K. Organization of Work at a Sheep Farm; OGIZ–Selkhozgiz: Moscow, Russia, 1947; 120 p.Matika, O.; Sechi, S.; Pong-Wong, R.; Houston, R.D.; Clop, A.; Woolliams, J.A.; Bishop, S.C. Characterization of OAR1 and OAR18 QTL associated with muscle depth in British commercial terminal sire sheep. Anim. Genet. 2011, 42, 172–180.
- All-Union Agricultural Exhibition of 1954; Ministry of Agriculture of the USSR: Moscow, Russia, 1955; 118 p.Mullen, M.P.; Hanrahan, J.P.; Howard, D.J.; Powell, R. Investigation of prolific sheep from UK and Ireland for evidence on origin of the mutations in BMP15 (FecXG, FecXB) and GDF9 (FecGH) in Belclare and Cambridge sheep. PLoS ONE 2013, 8, e53172.
- Ivanov, M.F. Sheep Farming. In Complete Works, in 7 Volumes; Greben, L.K., Ed.; Selkhozgiz: Moscow, Russia, 1964; Volume 4, 779 p.Stear, A.; Ali, A.O.A.; Brujeni, G.N.; Buitkamp, J.; Donskow-Łysoniewska, K.; Fairlie-Clarke, K.; Groth, D.; Isa, N.M.M.; Stear, M.J. Identification of the amino acids in the Major Histocompatibility Complex class II region of Scottish Blackface sheep that are associated with resistance to nematode infection. Int. J. Parasitol. 2019, 49, 797–804.
- Semyonov, S.I.; Selkin, I.I. Sheep. In Animal Genetic Resources of the USSR; Dmitriev, N.G., Ernst, L.K., Eds.; Food and Agriculture Organization of the United Nations: Rome, Italy, 1989; Volume 65, Chapter 4; pp. 154–271.Wilkie, H.; Riggio, V.; Matika, O.; Nicol, L.; Watt, K.A.; Sinclair, R.; Sparks, A.M.; Nussey, D.H.; Pemberton, J.M.; Houston, R.D.; et al. A candidate gene approach to study nematode resistance traits in naturally infected sheep. Vet. Parasitol. 2017, 243, 71–74.
- Kaneva, L.A.; Zharikov, Y.A.; Matyukov, V.S. Zootechnical characteristics of Pechora meat-wool semi-fine-fleece wool sheep. Agrarnaya Nauka Evro-Severo-Vostoka 2014, 5 (42), 58–63.Cinar, M.U.; Mousel, M.R.; Herndon, M.K.; Taylor, J.B.; White, S.N. Association of TMEM8B and SPAG8 with mature weight in sheep. Animals 2020, 10, 2391.
- Yearbook on Breeding Work in Sheep and Goat Farming in the Farms of the Russian Federation (2019); All-Russian Research Institute of Animal Breeding: Moscow, Russia, 2020; 344 p.Kijas, J.W.; Townley, D.; Dalrymple, B.P.; Heaton, M.P.; Maddox, J.F.; McGrath, A.; Wilson, P.; Ingersoll, R.G.; McCulloch, R.; McWilliam, S.; et al. A genome wide survey of SNP variation reveals the genetic structure of sheep breeds. PLoS ONE 2009, 4, e4668.
- Kuleshov, P.N. Value of Merino and English Meat Breeds in Improving Sheep Farming in the USSR; Moscow Higher Zootechnical Institute: Moscow, Russia, 1926; 16 p.International Sheep Genomics Consortium; Archibald, A.L.; Cockett, N.E.; Dalrymple, B.P.; Faraut, T.; Kijas, J.W.; Maddox, J.F.; McEwan, J.C.; Hutton Oddy, V.; Raadsma, H.W.; et al. The sheep genome reference sequence: A work in progress. Anim. Genet. 2010, 41, 449–453.
- Kuleshov, P.N. Meat-and-Wool Sheep Breeding; Selkhozgiz: Moscow, Russia, 1933; 112 p.Jiang, Y.; Xie, M.; Chen, W.; Talbot, R.; Maddox, J.F.; Faraut, T.; Wu, C.; Muzny, D.M.; Li, Y.; Zhang, W.; et al. The sheep genome illuminates biology of the rumen and lipid metabolism. Science 2014, 344, 1168–1173.
- Glembotsky, Y.L.; Deikhman, E.K.; Esaulov, P.A. Breeding in Sheep Farming: Achievements in Developing New Sheep Breeds and Improving Existing Ones; Selhozgiz: Moscow, Russia, 1946; 151 p.Worley, K.C.; English, A.C.; Richards, S.; Ross-Ibarra, J.; Han, Y.; Hughes, D.; Deiros, D.R.; Vee, V.; Wang, M.; Boerwinkle, E. Improving Genomes Using Long Reads and PBJelly 2, Proceedings of the International Plant and Animal Genome XXII Conference, San Diego, CA, USA, 10–15 January 2014; Scherago International: San Diego, CA, USA, 2014; Abstract P1033.
- Ostrovsky, A.V. Universal Reference Book on the History of Russia: With Tables, Diagrams and Dictionaries; Paritet: St. Petersburg, Russia, 2000; 384 p.National Center for Biotechnology Information, US National Library of Medicine. Genome Assembly: Oar_rambouillet_v1.0. Date: 2 November 2017. Available online: (accessed on 3 February 2021).
- Luschihina, E.M. Sheep breed resources of Kyrgyzstan. In Collection of Scientific Papers Based on the International Coordination Congress of Scientists Sheep Breeders 2013; Collection of Proceedings of SNIIZHK; Stavropol Research Institute of Animal Husbandry and Food Production: Stavropol, Russia, 2013; No. 6–1, pp. 67–80.National Center for Biotechnology Information, US National Library of Medicine. Genome Assembly: ASM1117029v1. Date: 11 March 2020. Available online: (accessed on 3 February 2021).
- Bowles, D. Recent advances in understanding the genetic resources of sheep breeds locally-adapted to the UK uplands: Opportunities they offer for sustainable productivity. Front. Genet. 2015, 6, 24.Mucha, S.; Bunger, L.; Conington, J. Genome-wide association study of footrot in Texel sheep. Genet. Sel. Evol. 2015, 47, 35.
- National Sheep Association. Sheep Breeds. Available online: https://www.nationalsheep.org.uk/uk-sheep-industry/sheep-inthe-uk/sheep-breeds/ (accessed on 3 February 2021).McEwan, J.; Dodds, K.; Rowe, S.; Brauning, R.; Clarke, S. Genomic Selection in Sheep: Where to Now? In Proceedings of the Book of Abstracts of the 67th Annual Meeting of the European Federation of Animal Science, Belfast, UK, 29 August–1 September 2016; Wageningen Academic Publishers: Wageningen, The Netherlands, 2016; Volume 22, p. 161.
- Boettcher, P.; Haile, A.; Hall, K.; Miller, J.L.; Mirkena, T.; Scherf, B.; Wurzinger, M. Animal genetic resources and adaptation. In The Second Report on the State of the World’s Animal Genetic Resources for Food and Agriculture; Scherf, B.D., Pilling, D., Eds.; FAO Commission on Genetic Resources for Food and Agriculture Assessments: Rome, Italy, 2015; p. 87.Beynon, S.E.; Slavov, G.T.; Farré, M.; Sunduimijid, B.; Waddams, K.; Davies, B.; Haresign, W.; Kijas, J.; MacLeod, I.M.; Newbold, C.J.; et al. Population structure and history of the Welsh sheep breeds determined by whole genome genotyping. BMC Genet. 2015, 16, 65.
- Baylis, M.; Chihota, C.; Stevenson, E.; Goldmann, W.; Smith, A.; Sivam, K.; Tongue, S.; Gravenor, M.B. Risk of scrapie in British sheep of different prion protein genotype. J. Gen. Virol. 2004, 85, 2735–2740.Daetwyler, H.D.; Brauning, R.; Chamberlain, A.J.; McWilliam, S.; McCulloch, A.; Vander Jagt, C.J.; Sunduimijid, B.; Hayes, B.J.; Kijas, J.W. 1000 Bull genomes and SheepGenomesDB projects: Enabling cost-effective sequence level analyses globally. In Proceedings of the 22nd Conference of the Association for the Advancement of Animal Breeding and Genetics (AAABG), Townsville, Australia, 2–5 July 2017; Association for the Advancement of Animal Breeding and Genetics: Armidale, Australia, 2017; Volume 22, pp. 201–204.
- Goldmann, W.; Baylis, M.; Chihota, C.; Stevenson, E.; Hunter, N. Frequencies of PrP gene haplotypes in British sheep flocks and the implications for breeding programmes. J. Appl. Microbiol. 2005, 98, 1294–1302.Clark, E.L.; Bush, S.J.; McCulloch, M.E.B.; Farquhar, I.L.; Young, R.; Lefevre, L.; Pridans, C.; Tsang, H.G.; Wu, C.; Afrasiabi, C.; et al. A high resolution atlas of gene expression in the domestic sheep (Ovis aries). PLoS Genet. 2017, 13, e1006997.
- Roden, J.A.; Nieuwhof, G.J.; Bishop, S.C.; Jones, D.A.; Haresign, W.; Gubbins, S. Breeding programmes for TSE resistance in British sheep. I. Assessing the impact on prion protein (PrP) genotype frequencies. Prev. Vet. Med. 2006, 73, 1–16.Dalrymple, B.P.; Oddy, V.H.; McEwan, J.C.; Kijas, J.W.; Xiang, R.; Bond, J.; Cockett, N.; Worley, K.; Smith, T.; Vercoe, P.E. From sheep SNP chips, genome sequences and transcriptomes via mechanisms to improved sheep breeding and management. In Proceedings of the 21st Conference of the Association for the Advancement of Animal Breeding and Genetics (AAABG), Lorne, Australia, 28–30 September 2015; Association for the Advancement of Animal Breeding and Genetics: Armidale, Australia, 2015; Volume 21, pp. 45–48.
- Tongue, S.C.; Pfeiffer, D.U.; Shearn, P.D.; Wilesmith, J.W. PrP genotype: A flock-level risk factor for scrapie? Prev. Vet. Med. 2009, 92, 309–323.Murdoch, B.M.; White, S.N.; Mousel, M.R.; Massa, A.T.; Worley, K.C.; Archibald, A.L.; Clark, E.L.; Dalrymple, B.; Kijas, J.W.; Clarke, S.; et al. The Ovine FAANG Project. In Proceedings of the International Plant and Animal Genome XXVI Conference, San Diego, CA, USA, 13–17 January 2018; Scherago International: San Diego, CA, USA, 2018. Abstract W618.
- Saunders, G.C.; Cawthraw, S.; Mountjoy, S.J.; Hope, J.; Windl, O. PrP genotypes of atypical scrapie cases in Great Britain. J. Gen. Virol. 2006, 87, 3141–3149.Andersson, L.; Archibald, A.L.; Bottema, C.D.; Brauning, R.; Burgess, S.C.; Burt, D.W.; Casas, E.; Cheng, H.H.; Clarke, L.; Couldrey, C.; et al. Coordinated international action to accelerate genome-to-phenome with FAANG, the Functional Annotation of Animal Genomes project. Genome Biol. 2015, 16, 57.
- Freking, B.A.; Murphy, S.K.; Wylie, A.A.; Rhodes, S.J.; Keele, J.W.; Leymaster, K.A.; Jirtle, R.L.; Smith, T.P. Identification of the single base change causing the callipyge muscle hypertrophy phenotype, the only known example of polar overdominance in mammals. Genome Res. 2002, 12, 1496–1506.Naval-Sanchez, M.; Nguyen, Q.; McWilliam, S.; Porto-Neto, L.R.; Tellam, R.; Vuocolo, T.; Reverter, A.; Perez-Enciso, M.; Brauning, R.; Clarke, S.; et al. Sheep genome functional annotation reveals proximal regulatory elements contributed to the evolution of modern breeds. Nat. Commun. 2018, 9, 859.
- Zamorano, M.J.; Ruiter, J.; Townsend, S.; Cruickshank, R.; Bruford, M.; Byrne, K.; Rodero, A.; Vega-Pla, J.L. Polimorfismos de DNA en las razas ovinas Merino y Churra lebrijana: DNA polymorphisms in Merino and Churra lebrijana Sheep breeds. Arch. Zoot. 1998, 47, 267–272.Pariset, L.; Cappuccio, I.; Ajmone-Marsan, P.; Bruford, M.; Dunner, S.; Cortes, O.; Erhardt, G.; Prinzenberg, E.M.; Gutscher, K.; Joost, S.; et al. Characterization of 37 breed-specific single-nucleotide polymorphisms in sheep. J. Hered. 2006, 97, 531–534.
- Peter, C.; Bruford, M.; Perez, T.; Dalamitra, S.; Hewitt, G.; Erhardt, G.; Econogene Consortium. Genetic diversity and subdivision of 57 European and Middle-Eastern sheep breeds. Anim. Genet. 2007, 38, 37–44.Alberto, F.J.; Boyer, F.; Orozco-terWengel, P.; Streeter, I.; Servin, B.; de Villemereuil, P.; Benjelloun, B.; Librado, P.; Biscarini, F.; Colli, L.; et al. Convergent genomic signatures of domestication in sheep and goats. Nat. Commun. 2018, 9, 813.
- Lawson Handley, L.J.; Byrne, K.; Santucci, F.; Townsend, S.; Taylor, M.; Bruford, M.W.; Hewitt, G.M. Genetic structure of European sheep breeds. Heredity 2007, 99, 620–631.Deniskova, T.E.; Dotsev, A.V.; Selionova, M.I.; Kunz, E.; Medugorac, I.; Reyer, H.; Wimmers, K.; Barbato, M.; Traspov, A.A.; Brem, G.; et al. Population structure and genetic diversity of twenty-five Russian sheep breeds based on whole-genome genotyping. Genet. Sel. Evol. 2018, 50, 29.
- Russo-Almeida, P.A. Diversidade Genética e Diferenciação das Raças Portuguesas de Ovinos com Base em Marcadores de DNA–Microssatélites: Uma Perspectiva de Conservação. Tese de Doutorado em Ciência Animal, Universidade de Trás-os-Montes e Alto Douro, Vila Real, Portugal, 2007.Deniskova, T.; Dotsev, A.; Lushihina, E.; Shakhin, A.; Kunz, E.; Medugorac, I.; Reyer, H.; Wimmers, K.; Khayatzadeh, N.; Sölkner, J.; et al. Population structure and genetic diversity of sheep breeds in the Kyrgyzstan. Front. Genet. 2019, 10, 1311.
- Bowles, D.; Carson, A.; Isaac, P. Genetic distinctiveness of the Herdwick sheep breed and two other locally adapted hill breeds of the UK. PLoS ONE 2014, 9, e87823.Barbato, M.; Hailer, F.; Orozco-terWengel, P.; Kijas, J.; Mereu, P.; Cabras, P.; Mazza, R.; Pirastru, M.; Bruford, M.W. Genomic signatures of adaptive introgression from European mouflon into domestic sheep. Sci. Rep. 2017, 7, 7623.
- Bruford, M.; Townsend, S.J. Mitochondrial DNA diversity in modern sheep: Implications for domestication. In Documenting Domestication: New Genetic and Archaeological Paradigms; Zeder, M.A., Bradley, D.G., Emshwiller, E., Smith, B.D., Eds.; University of California Press: Berkeley, CA, USA, 2006; pp. 307–317.Kijas, J.W.; Lenstra, J.A.; Hayes, B.; Boitard, S.; Porto Neto, L.R.; San Cristobal, M.; Servin, B.; McCulloch, R.; Whan, V.; Gietzen, K.; et al. Genome-wide analysis of the world’s sheep breeds reveals high levels of historic mixture and strong recent selection. PLoS Biol. 2012, 10, e1001258.
- Pariset, L.; Mariotti, M.; Gargani, M.; Joost, S.; Negrini, R.; Perez, T.; Bruford, M.; Ajmone Marsan, P.; Valentini, A. Genetic diversity of sheep breeds from Albania, Greece, and Italy assessed by mitochondrial DNA and nuclear polymorphisms (SNPs). Sci. World J. 2011, 11, 1641–1659.Kijas, J. ISGC SNP50 HapMap and Sheep Breed Diversity Genotypes. v1. CSIRO. Data Collection. Published 6 May 2013. Available online: (accessed on 3 February 2021).
- Lv, F.H.; Peng, W.F.; Yang, J.; Zhao, Y.X.; Li, W.R.; Liu, M.J.; Ma, Y.H.; Zhao, Q.J.; Yang, G.L.; Wang, F.; et al. Mitogenomic meta-analysis identifies two phases of migration in the history of Eastern Eurasian sheep. Mol. Biol. Evol. 2015, 32, 2515–2533.Chen, Z.H.; Zhang, M.; Lv, F.H.; Ren, X.; Li, W.R.; Liu, M.J.; Nam, K.; Bruford, M.W.; Li, M.H. Contrasting patterns of genomic diversity reveal accelerated genetic drift but reduced directional selection on X-chromosome in wild and domestic sheep species. Genome Biol. Evol. 2018, 10, 1282–1297.
- Chessa, B.; Pereira, F.; Arnaud, F.; Amorim, A.; Goyache, F.; Mainland, I.; Kao, R.R.; Pemberton, J.M.; Beraldi, D.; Stear, M.J.; et al. Revealing the history of sheep domestication using retrovirus integrations. Science 2009, 324, 532–536.Davenport, K.M.; Hiemke, C.; McKay, S.D.; Thorne, J.W.; Lewis, R.M.; Taylor, T.; Murdoch, B.M. Genetic structure and admixture in sheep from terminal breeds in the United States. Anim. Genet. 2020, 51, 284–291.
- Matika, O.; Sechi, S.; Pong-Wong, R.; Houston, R.D.; Clop, A.; Woolliams, J.A.; Bishop, S.C. Characterization of OAR1 and OAR18 QTL associated with muscle depth in British commercial terminal sire sheep. Anim. Genet. 2011, 42, 172–180.Zhang, L.; Mousel, M.R.; Wu, X.; Michal, J.J.; Zhou, X.; Ding, B.; Dodson, M.V.; El-Halawany, N.K.; Lewis, G.S.; Jiang, Z. Genome-wide genetic diversity and differentially selected regions among Suffolk, Rambouillet, Columbia, Polypay, and Targhee sheep. PLoS ONE 2013, 8, e65942.
- Mullen, M.P.; Hanrahan, J.P.; Howard, D.J.; Powell, R. Investigation of prolific sheep from UK and Ireland for evidence on origin of the mutations in BMP15 (FecXG, FecXB) and GDF9 (FecGH) in Belclare and Cambridge sheep. PLoS ONE 2013, 8, e53172.Gutiérrez-Gil, B.; Arranz, J.J.; Pong-Wong, R.; García-Gámez, E.; Kijas, J.; Wiener, P. Application of selection mapping to identify genomic regions associated with dairy production in sheep. PLoS ONE 2014, 9, e94623.
- Stear, A.; Ali, A.O.A.; Brujeni, G.N.; Buitkamp, J.; Donskow-Łysoniewska, K.; Fairlie-Clarke, K.; Groth, D.; Isa, N.M.M.; Stear, M.J. Identification of the amino acids in the Major Histocompatibility Complex class II region of Scottish Blackface sheep that are associated with resistance to nematode infection. Int. J. Parasitol. 2019, 49, 797–804.Gutiérrez-Gil, B.; Esteban-Blanco, C.; Suarez-Vega, A.; Arranz, J.J. Detection of quantitative trait loci and putative causal variants affecting somatic cell score in dairy sheep by using a 50K SNP-Chip and whole genome sequencing. J. Dairy Sci. 2018, 101, 9072–9088.
- Wilkie, H.; Riggio, V.; Matika, O.; Nicol, L.; Watt, K.A.; Sinclair, R.; Sparks, A.M.; Nussey, D.H.; Pemberton, J.M.; Houston, R.D.; et al. A candidate gene approach to study nematode resistance traits in naturally infected sheep. Vet. Parasitol. 2017, 243, 71–74.Atlija, M.; Arranz, J.J.; Martinez-Valladares, M.; Gutiérrez-Gil, B. Detection and replication of QTL underlying resistance to gastrointestinal nematodes in adult sheep using the ovine 50K SNP array. Genet. Sel. Evol. 2016, 48, 4.
- Cinar, M.U.; Mousel, M.R.; Herndon, M.K.; Taylor, J.B.; White, S.N. Association of TMEM8B and SPAG8 with mature weight in sheep. Animals 2020, 10, 2391.Banos, G.; Bramis, G.; Bush, S.J.; Clark, E.L.; McCulloch, M.E.B.; Smith, J.; Schulze, G.; Arsenos, G.; Hume, D.A.; Psifidi, A. The genomic architecture of mastitis resistance in dairy sheep. BMC Genom. 2017, 18, 624.
- Kijas, J.W.; Townley, D.; Dalrymple, B.P.; Heaton, M.P.; Maddox, J.F.; McGrath, A.; Wilson, P.; Ingersoll, R.G.; McCulloch, R.; McWilliam, S.; et al. A genome wide survey of SNP variation reveals the genetic structure of sheep breeds. PLoS ONE 2009, 4, e4668.Sheep QTLdb. Animal QTLdb, NAGRP—Bioinformatics Coordination Program. Available online: (accessed on 3 February 2021).
- International Sheep Genomics Consortium; Archibald, A.L.; Cockett, N.E.; Dalrymple, B.P.; Faraut, T.; Kijas, J.W.; Maddox, J.F.; McEwan, J.C.; Hutton Oddy, V.; Raadsma, H.W.; et al. The sheep genome reference sequence: A work in progress. Anim. Genet. 2010, 41, 449–453.Riggio, V.; Matika, O.; Pong-Wong, R.; Stear, M.J.; Bishop, S.C. Genome-wide association and regional heritability mapping to identify loci underlying variation in nematode resistance and body weight in Scottish Blackface lambs. Heredity 2013, 110, 420–429.
- Jiang, Y.; Xie, M.; Chen, W.; Talbot, R.; Maddox, J.F.; Faraut, T.; Wu, C.; Muzny, D.M.; Li, Y.; Zhang, W.; et al. The sheep genome illuminates biology of the rumen and lipid metabolism. Science 2014, 344, 1168–1173.Riggio, V.; Pong-Wong, R.; Sallé, G.; Usai, M.G.; Casu, S.; Moreno, C.R.; Matika, O.; Bishop, S.C. A joint analysis to identify loci underlying variation in nematode resistance in three European sheep populations. J. Anim. Breed. Genet. 2014, 131, 426–436.
- Worley, K.C.; English, A.C.; Richards, S.; Ross-Ibarra, J.; Han, Y.; Hughes, D.; Deiros, D.R.; Vee, V.; Wang, M.; Boerwinkle, E. Improving Genomes Using Long Reads and PBJelly 2, Proceedings of the International Plant and Animal Genome XXII Conference, San Diego, CA, USA, 10–15 January 2014; Scherago International: San Diego, CA, USA, 2014; Abstract P1033.Riggio, V.; Abdel-Aziz, M.; Matika, O.; Moreno, C.R.; Carta, A.; Bishop, S.C. Accuracy of genomic prediction within and across populations for nematode resistance and body weight traits in sheep. Animal 2014, 8, 520–528.
- National Center for Biotechnology Information, US National Library of Medicine. Genome Assembly: Oar_rambouillet_v1.0. Date: 2 November 2017. Available online: https://www.ncbi.nlm.nih.gov/assembly/GCA_002742125.1 (accessed on 3 February 2021).Keane, O.M.; Hanrahan, J.P.; McRae, K.M.; Good, B. An independent validation study of loci associated with nematode resistance in sheep. Anim. Genet. 2018, 49, 265–268.
- National Center for Biotechnology Information, US National Library of Medicine. Genome Assembly: ASM1117029v1. Date: 11 March 2020. Available online: https://www.ncbi.nlm.nih.gov/assembly/GCA_011170295.1/ (accessed on 3 February 2021).Purfield, D.C.; McParland, S.; Wall, E.; Berry, D.P. The distribution of runs of homozygosity and selection signatures in six commercial meat sheep breeds. PLoS ONE 2017, 12, e0176780.
- McEwan, J.; Dodds, K.; Rowe, S.; Brauning, R.; Clarke, S. Genomic Selection in Sheep: Where to Now? In Proceedings of the Book of Abstracts of the 67th Annual Meeting of the European Federation of Animal Science, Belfast, UK, 29 August–1 September 2016; Wageningen Academic Publishers: Wageningen, The Netherlands, 2016; Volume 22, p. 161.Yang, J.; Li, W.R.; Lv, F.H.; He, S.G.; Tian, S.L.; Peng, W.F.; Sun, Y.W.; Zhao, Y.X.; Tu, X.L.; Zhang, M.; et al. Whole-genome sequencing of native sheep provides insights into rapid adaptations to extreme environments. Mol. Biol. Evol. 2016, 33, 2576–2592.
- Daetwyler, H.D.; Brauning, R.; Chamberlain, A.J.; McWilliam, S.; McCulloch, A.; Vander Jagt, C.J.; Sunduimijid, B.; Hayes, B.J.; Kijas, J.W. 1000 Bull genomes and SheepGenomesDB projects: Enabling cost-effective sequence level analyses globally. In Proceedings of the 22nd Conference of the Association for the Advancement of Animal Breeding and Genetics (AAABG), Townsville, Australia, 2–5 July 2017; Association for the Advancement of Animal Breeding and Genetics: Armidale, Australia, 2017; Volume 22, pp. 201–204.Nosrati, M.; Asadollahpour Nanaei, H.; Amiri Ghanatsaman, Z.; Esmailizadeh, A. Whole genome sequence analysis to detect signatures of positive selection for high fecundity in sheep. Reprod. Domest. Anim. 2019, 54, 358–364.
- Clark, E.L.; Bush, S.J.; McCulloch, M.E.B.; Farquhar, I.L.; Young, R.; Lefevre, L.; Pridans, C.; Tsang, H.G.; Wu, C.; Afrasiabi, C.; et al. A high resolution atlas of gene expression in the domestic sheep (Ovis aries). PLoS Genet. 2017, 13, e1006997.Zhang, Y.; Xue, X.; Liu, Y.; Abied, A.; Ding, Y.; Zhao, S.; Wang, W.; Ma, L.; Guo, J.; Guan, W.; et al. Genome-wide comparative analyses reveal selection signatures underlying adaptation and production in Tibetan and Poll Dorset sheep. Sci. Rep. 2021, 11, 2466.
- Dalrymple, B.P.; Oddy, V.H.; McEwan, J.C.; Kijas, J.W.; Xiang, R.; Bond, J.; Cockett, N.; Worley, K.; Smith, T.; Vercoe, P.E. From sheep SNP chips, genome sequences and transcriptomes via mechanisms to improved sheep breeding and management. In Proceedings of the 21st Conference of the Association for the Advancement of Animal Breeding and Genetics (AAABG), Lorne, Australia, 28–30 September 2015; Association for the Advancement of Animal Breeding and Genetics: Armidale, Australia, 2015; Volume 21, pp. 45–48.National Center for Biotechnology Information, US National Library of Medicine. BioProject: Search Results. Available online: (accessed on 3 February 2021).
- Murdoch, B.M.; White, S.N.; Mousel, M.R.; Massa, A.T.; Worley, K.C.; Archibald, A.L.; Clark, E.L.; Dalrymple, B.; Kijas, J.W.; Clarke, S.; et al. The Ovine FAANG Project. In Proceedings of the International Plant and Animal Genome XXVI Conference, San Diego, CA, USA, 13–17 January 2018; Scherago International: San Diego, CA, USA, 2018. Abstract W618.EMBL-EBI. The European Nucleotide Archive (ENA) Browser. Project: PRJEB14685. Available online: (accessed on 3 February 2021).
- Andersson, L.; Archibald, A.L.; Bottema, C.D.; Brauning, R.; Burgess, S.C.; Burt, D.W.; Casas, E.; Cheng, H.H.; Clarke, L.; Couldrey, C.; et al. Coordinated international action to accelerate genome-to-phenome with FAANG, the Functional Annotation of Animal Genomes project. Genome Biol. 2015, 16, 57.Li, X.; Yang, J.; Shen, M.; Xie, X.L.; Liu, G.J.; Xu, Y.X.; Lv, F.H.; Yang, H.; Yang, Y.L.; Liu, C.B.; et al. Whole-genome resequencing of wild and domestic sheep identifies genes associated with morphological and agronomic traits. Nat. Commun. 2020, 11, 2815.
- Naval-Sanchez, M.; Nguyen, Q.; McWilliam, S.; Porto-Neto, L.R.; Tellam, R.; Vuocolo, T.; Reverter, A.; Perez-Enciso, M.; Brauning, R.; Clarke, S.; et al. Sheep genome functional annotation reveals proximal regulatory elements contributed to the evolution of modern breeds. Nat. Commun. 2018, 9, 859.
- Pariset, L.; Cappuccio, I.; Ajmone-Marsan, P.; Bruford, M.; Dunner, S.; Cortes, O.; Erhardt, G.; Prinzenberg, E.M.; Gutscher, K.; Joost, S.; et al. Characterization of 37 breed-specific single-nucleotide polymorphisms in sheep. J. Hered. 2006, 97, 531–534.
- Alberto, F.J.; Boyer, F.; Orozco-terWengel, P.; Streeter, I.; Servin, B.; de Villemereuil, P.; Benjelloun, B.; Librado, P.; Biscarini, F.; Colli, L.; et al. Convergent genomic signatures of domestication in sheep and goats. Nat. Commun. 2018, 9, 813.
- Deniskova, T.E.; Dotsev, A.V.; Selionova, M.I.; Kunz, E.; Medugorac, I.; Reyer, H.; Wimmers, K.; Barbato, M.; Traspov, A.A.; Brem, G.; et al. Population structure and genetic diversity of twenty-five Russian sheep breeds based on whole-genome genotyping. Genet. Sel. Evol. 2018, 50, 29.
- Deniskova, T.; Dotsev, A.; Lushihina, E.; Shakhin, A.; Kunz, E.; Medugorac, I.; Reyer, H.; Wimmers, K.; Khayatzadeh, N.; Sölkner, J.; et al. Population structure and genetic diversity of sheep breeds in the Kyrgyzstan. Front. Genet. 2019, 10, 1311.
- Barbato, M.; Hailer, F.; Orozco-terWengel, P.; Kijas, J.; Mereu, P.; Cabras, P.; Mazza, R.; Pirastru, M.; Bruford, M.W. Genomic signatures of adaptive introgression from European mouflon into domestic sheep. Sci. Rep. 2017, 7, 7623.
- Kijas, J.W.; Lenstra, J.A.; Hayes, B.; Boitard, S.; Porto Neto, L.R.; San Cristobal, M.; Servin, B.; McCulloch, R.; Whan, V.; Gietzen, K.; et al. Genome-wide analysis of the world’s sheep breeds reveals high levels of historic mixture and strong recent selection. PLoS Biol. 2012, 10, e1001258.
- Kijas, J. ISGC SNP50 HapMap and Sheep Breed Diversity Genotypes. v1. CSIRO. Data Collection. Published 6 May 2013. Available online: https://data.csiro.au/collections/collection/CIcsiro:6494v1 (accessed on 3 February 2021).
- Chen, Z.H.; Zhang, M.; Lv, F.H.; Ren, X.; Li, W.R.; Liu, M.J.; Nam, K.; Bruford, M.W.; Li, M.H. Contrasting patterns of genomic diversity reveal accelerated genetic drift but reduced directional selection on X-chromosome in wild and domestic sheep species. Genome Biol. Evol. 2018, 10, 1282–1297.
- Davenport, K.M.; Hiemke, C.; McKay, S.D.; Thorne, J.W.; Lewis, R.M.; Taylor, T.; Murdoch, B.M. Genetic structure and admixture in sheep from terminal breeds in the United States. Anim. Genet. 2020, 51, 284–291.
- Zhang, L.; Mousel, M.R.; Wu, X.; Michal, J.J.; Zhou, X.; Ding, B.; Dodson, M.V.; El-Halawany, N.K.; Lewis, G.S.; Jiang, Z. Genome-wide genetic diversity and differentially selected regions among Suffolk, Rambouillet, Columbia, Polypay, and Targhee sheep. PLoS ONE 2013, 8, e65942.
- Gutiérrez-Gil, B.; Arranz, J.J.; Pong-Wong, R.; García-Gámez, E.; Kijas, J.; Wiener, P. Application of selection mapping to identify genomic regions associated with dairy production in sheep. PLoS ONE 2014, 9, e94623.
- Gutiérrez-Gil, B.; Esteban-Blanco, C.; Suarez-Vega, A.; Arranz, J.J. Detection of quantitative trait loci and putative causal variants affecting somatic cell score in dairy sheep by using a 50K SNP-Chip and whole genome sequencing. J. Dairy Sci. 2018, 101, 9072–9088.
- Atlija, M.; Arranz, J.J.; Martinez-Valladares, M.; Gutiérrez-Gil, B. Detection and replication of QTL underlying resistance to gastrointestinal nematodes in adult sheep using the ovine 50K SNP array. Genet. Sel. Evol. 2016, 48, 4.
- Banos, G.; Bramis, G.; Bush, S.J.; Clark, E.L.; McCulloch, M.E.B.; Smith, J.; Schulze, G.; Arsenos, G.; Hume, D.A.; Psifidi, A. The genomic architecture of mastitis resistance in dairy sheep. BMC Genom. 2017, 18, 624.
- Sheep QTLdb. Animal QTLdb, NAGRP—Bioinformatics Coordination Program. Available online: https://www.animalgenome.org/cgi-bin/QTLdb/OA/index (accessed on 3 February 2021).
- Riggio, V.; Matika, O.; Pong-Wong, R.; Stear, M.J.; Bishop, S.C. Genome-wide association and regional heritability mapping to identify loci underlying variation in nematode resistance and body weight in Scottish Blackface lambs. Heredity 2013, 110, 420–429.
- Riggio, V.; Pong-Wong, R.; Sallé, G.; Usai, M.G.; Casu, S.; Moreno, C.R.; Matika, O.; Bishop, S.C. A joint analysis to identify loci underlying variation in nematode resistance in three European sheep populations. J. Anim. Breed. Genet. 2014, 131, 426–436.
- Riggio, V.; Abdel-Aziz, M.; Matika, O.; Moreno, C.R.; Carta, A.; Bishop, S.C. Accuracy of genomic prediction within and across populations for nematode resistance and body weight traits in sheep. Animal 2014, 8, 520–528.
- Keane, O.M.; Hanrahan, J.P.; McRae, K.M.; Good, B. An independent validation study of loci associated with nematode resistance in sheep. Anim. Genet. 2018, 49, 265–268.
- Purfield, D.C.; McParland, S.; Wall, E.; Berry, D.P. The distribution of runs of homozygosity and selection signatures in six commercial meat sheep breeds. PLoS ONE 2017, 12, e0176780.
- Yang, J.; Li, W.R.; Lv, F.H.; He, S.G.; Tian, S.L.; Peng, W.F.; Sun, Y.W.; Zhao, Y.X.; Tu, X.L.; Zhang, M.; et al. Whole-genome sequencing of native sheep provides insights into rapid adaptations to extreme environments. Mol. Biol. Evol. 2016, 33, 2576–2592.
- Nosrati, M.; Asadollahpour Nanaei, H.; Amiri Ghanatsaman, Z.; Esmailizadeh, A. Whole genome sequence analysis to detect signatures of positive selection for high fecundity in sheep. Reprod. Domest. Anim. 2019, 54, 358–364.
- Zhang, Y.; Xue, X.; Liu, Y.; Abied, A.; Ding, Y.; Zhao, S.; Wang, W.; Ma, L.; Guo, J.; Guan, W.; et al. Genome-wide comparative analyses reveal selection signatures underlying adaptation and production in Tibetan and Poll Dorset sheep. Sci. Rep. 2021, 11, 2466.
- National Center for Biotechnology Information, US National Library of Medicine. BioProject: Search Results. Available online: https://www.ncbi.nlm.nih.gov/bioproject?term=10709[top+bioproject]+NOT+160933[uid] (accessed on 3 February 2021).
- EMBL-EBI. The European Nucleotide Archive (ENA) Browser. Project: PRJEB14685. Available online: https://www.ebi.ac.uk/ena/data/view/PRJEB14685 (accessed on 3 February 2021).
- Li, X.; Yang, J.; Shen, M.; Xie, X.L.; Liu, G.J.; Xu, Y.X.; Lv, F.H.; Yang, H.; Yang, Y.L.; Liu, C.B.; et al. Whole-genome resequencing of wild and domestic sheep identifies genes associated with morphological and agronomic traits. Nat. Commun. 2020, 11, 2815.
