Tamoxifen is an important adjuvant endocrine therapy in estrogen receptor (ER)-positive breast cancer patients. It is metabolized into its most active antiestrogenic metabolite endoxifen, predominantly by cytochrome P450 2D6 (CYP2D6). Many factors, including genetic variation in CYP2D6, influence tamoxifen metabolism and pharmacokinetics.
- tamoxifen
- Pharmacogenetics
- PGx
- CYP2D6
- Cytochrome P450 2D6
- endoxifen
- tamoxifen metabolism
- tamoxifen pharmacokinetics
1. Introduction
For many years, tamoxifen has been known as the most important adjuvant endocrine treatment in patients with estrogen receptor (ER) positive breast cancer [1,2]. It is a selective estrogen receptor modulator (SERM), which inhibits tumor growth and promotes apoptosis in ER-positive tumors [3], resulting in a reduced risk of recurrence and death from breast cancer [4]. Tamoxifen is metabolized into its most active antiestrogenic metabolite endoxifen, predominantly by cytochrome P450 2D6 (CYP2D6). Despite high efficacy, a wide variability in the response of individuals to standard doses of tamoxifen is seen [4]. Factors influencing drug responses such as gender, age, obesity, drug–drug interactions, drug–food interactions, treatment adherence, comorbidity, liver and renal function, pregnancy, circadian rhythm, as well as genetic factors could possibly explain this wide variability [5–10].
For many years, tamoxifen has been known as the most important adjuvant endocrine treatment in patients with estrogen receptor (ER) positive breast cancer [1][2]. It is a selective estrogen receptor modulator (SERM), which inhibits tumor growth and promotes apoptosis in ER-positive tumors [3], resulting in a reduced risk of recurrence and death from breast cancer [4]. Tamoxifen is metabolized into its most active antiestrogenic metabolite endoxifen, predominantly by cytochrome P450 2D6 (CYP2D6). Despite high efficacy, a wide variability in the response of individuals to standard doses of tamoxifen is seen [4]. Factors influencing drug responses such as gender, age, obesity, drug–drug interactions, drug–food interactions, comorbidity, liver and renal function, pregnancy, circadian rhythm, as well as genetic factors could possibly explain this wide variability [5][6][7][8][9][10].
As genetic variation in CYP2D6 leads to altered enzyme activity and thereby potentially to an affected efficacy [5], pharmacogenetic (PGx) testing could play a major role in optimizing tamoxifen treatment. Nowadays, PGx testing is available for an increasing number of drugs, mainly in psychiatric but also for cardiologic and oncologic applications. However, only a small number of drugs are considered to require upfront genotyping, such as HLA-B*5701 genotyping for abacavir, CYP2C19 genotyping for clopidogrel, and DPYD testing for fluoropyrimidines treatment [11–13] The challenge of PGx testing is to obtain actionable information of genetic variants and their influence on outcome. Experts differ in their interpretation of published evidence and their recommendations [5]. This also applies to tamoxifen. Whereas the first studies on CYP2D6 genotyping for optimizing tamoxifen therapy based on pharmacokinetic studies [14] or outcome [15,16] were published in 2005, its clinical implementation is still being debated [17,18]. The controversy was most prominently seen in 2012. At that time, two studies were published, and both concluded there was no significant association between CYP2D6 genotype and outcome in breast cancer patients treated with adjuvant tamoxifen therapy [19,20]. This triggered several researchers with opposite visions, leading to a correspondence as published in the JNCI [21–24]. In 2013, several meta-analyses were performed in order to answer the question of whether CYP2D6 status affects clinical outcomes in tamoxifen therapy [25–29]. However, also these findings yielded contradictory results. In 2018, the Clinical Pharmacogenetics Implementation Consortium (CPIC) published a recommendation on CYP2D6 genotyping for guiding adjuvant tamoxifen therapy, thus supporting the importance of the CYP2D6 genotype in tamoxifen therapy. However, the European Society for Medical Oncology (ESMO) indicated in a publication in 2019 that according to their view, there was no place for CYP2D6 genotyping in a clinical setting [18].
As genetic variation in CYP2D6 leads to altered enzyme activity and thereby potentially to an affected efficacy [5], pharmacogenetic (PGx) testing could play a major role in optimizing tamoxifen treatment. Nowadays, PGx testing is available for an increasing number of drugs, mainly in psychiatric but also for cardiologic and oncologic applications. However, only a small number of drugs are considered to require upfront genotyping, such as HLA-B*5701 genotyping for abacavir, CYP2C19 genotyping for clopidogrel, and DPYD testing for fluoropyrimidines treatment [11][12][13]. The challenge of PGx testing is to obtain actionable information of genetic variants and their influence on outcome. Experts differ in their interpretation of published evidence and their recommendations [5]. This also applies to tamoxifen. Whereas the first studies on CYP2D6 genotyping for optimizing tamoxifen therapy based on pharmacokinetic studies [14] or outcome [15][16] were published in 2005, its clinical implementation is still being debated [17][18]. The controversy was most prominently seen in 2012. At that time, two studies were published, and both concluded there was no significant association between CYP2D6 genotype and outcome in breast cancer patients treated with adjuvant tamoxifen therapy [19][20]. This triggered several researchers with opposite visions, leading to a correspondence as published in the JNCI [21][22][23][24]. In 2013, several meta-analyses were performed in order to answer the question of whether CYP2D6 status affects clinical outcomes in tamoxifen therapy [25][26][27][28][29]. However, also these findings yielded contradictory results. In 2018, the Clinical Pharmacogenetics Implementation Consortium (CPIC) published a recommendation on CYP2D6 genotyping for guiding adjuvant tamoxifen therapy, thus supporting the importance of the CYP2D6 genotype in tamoxifen therapy. However, the European Society for Medical Oncology (ESMO) indicated in a publication in 2019 that according to their view, there was no place for CYP2D6 genotyping in a clinical setting [18].
According to predictions, over 3 million women will be diagnosed with breast cancer in 2040 [30], which triggers the strong need for optimizing tamoxifen treatment.
2. Tamoxifen Metabolism
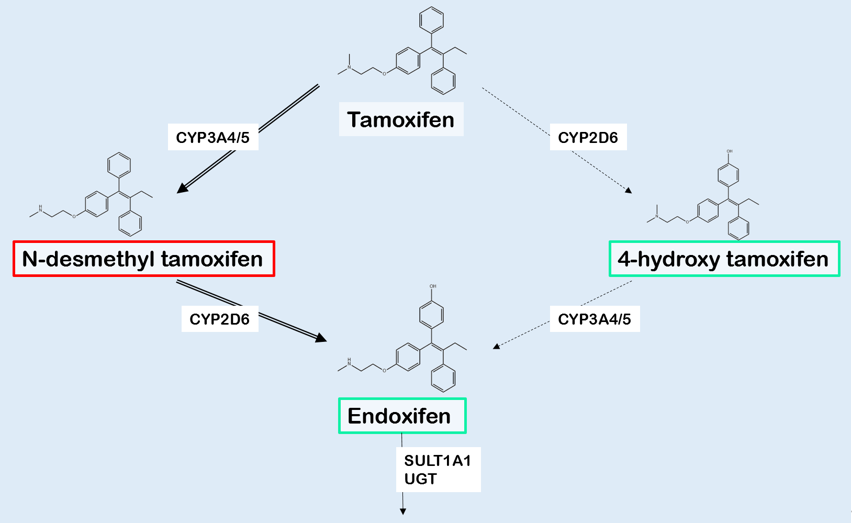
Tamoxifen is a prodrug, which is converted into multiple derivatives by phase I enzymes. Among these, 4-hydroxytamoxifen (4OH-TAM) and endoxifen show the strongest affinity with the ER [2,31][2][31]. As the plasma concentration of endoxifen is 6 to 12 times higher compared with 4OH-TAM, and endoxifen has the lowest IC50 at the ER, endoxifen is considered the major active tamoxifen metabolite [2,31][2][31]. Tamoxifen is N-demethylated, predominantly by CYP3A4 and CYP3A5, resulting in inactive N-desmethyltamoxifen (NDM-TAM) (Figure 1). 4-hydroxylation of NDM-TAM is nearly exclusively performed by CYP2D6, resulting in the formation of 4-hydroxy-N-desmethyltamoxifen, also known as endoxifen [2,31][2][31]. As shown in Figure 1, demethylation and hydroxylation can also occur in the opposite order, resulting a different intermediate metabolite called 4-hydroxytamoxifen (4OH-TAM) [2,31][2][31]. As NDM-TAM is the primary metabolite regarding plasma concentrations, the demethylation followed by hydroxylation is supposed to be the main route [32].
Figure 1. Simplistic representation of the main biotransformation of tamoxifen and its metabolites. The generation of N-desmethyltamoxifen (NDM-TAM) is predominantly catalyzed by CYP3A4/5, whereas especially CYP2D6 is responsible for the formation of 4-hydroxytamoxifen (4OH-TAM) and endoxifen. The activity of metabolites is shown using red to indicate for inactivity and green for activity. The various metabolites are inactivated by UGTs and SULTs, mainly isoform SULT1A1. Abbreviations: CYP: Cytochrome P450 isoenzymes, UGT: UDP-glucuronosyltransferase, SULT: sulfotransferase isoenzyme. Selfmade fFigure, based on Jin, et al. [14].
3. Cytochrome P450 2D6
CYP2D6 is involved in the metabolism of ≈25% of the most commonly used drugs, whereas it only accounts for approximately 2% of total liver CYP protein capacity [2,33]. As shown in Figure 1, CYP2D6 is responsible for the specific conversion of NDM-TAM to endoxifen [31]. Therefore, CYP2D6 is considered the most important drug-metabolizing enzyme in tamoxifen metabolism. The CYP2D6 gene is located on chromosome 22q13.2 and is highly polymorphic [34]. Currently, approximately 150 single nucleotide polymorphisms (SNPs) and 100 allelic variants are described [35]. Genetic polymorphisms may result in non-functional or reduced function alleles. Copy number variations, such as CYP2D6 gene deletions and CYP2D6 gene duplications, also occur [31,36]. This genetic variability results in individuals showing a broad spectrum of enzyme activities, indicated as poor (PM), intermediate (IM), normal (NM) or ultra-rapid metabolizers (UM) based on genetic composition [17,36]. Another approach is to use Activity Scores (AS), in which normal alleles are assigned a value of 1.0, decreased activity alleles are assigned a score of 0.5, and null alleles are assigned a score of 0.0 [17]. Genetic variants can be specific for certain populations and rarely found in other populations [33,37]. The variation in allele frequency results in differences in metabolic CYP2D6 activity amongst ethnic groups [33].
CYP2D6 is involved in the metabolism of ≈25% of the most commonly used drugs, whereas it only accounts for approximately 2% of total liver CYP protein capacity [2][33]. As shown in Figure 1, CYP2D6 is responsible for the specific conversion of NDM-TAM to endoxifen [31]. Therefore, CYP2D6 is considered the most important drug-metabolizing enzyme in tamoxifen metabolism. The CYP2D6 gene is located on chromosome 22q13.2 and is highly polymorphic [34]. Currently, approximately 150 single nucleotide polymorphisms (SNPs) and 100 allelic variants are described [35]. Genetic polymorphisms may result in non-functional or reduced function alleles. Copy number variations, such as CYP2D6 gene deletions and CYP2D6 gene duplications, also occur [31][36]. This genetic variability results in individuals showing a broad spectrum of enzyme activities, indicated as poor (PM), intermediate (IM), normal (NM) or ultra-rapid metabolizers (UM) based on genetic composition [17][36]. Another approach is to use Activity Scores (AS), in which normal alleles are assigned a value of 1.0, decreased activity alleles are assigned a score of 0.5, and null alleles are assigned a score of 0.0 [17]. Genetic variants can be specific for certain populations and rarely found in other populations [33][37]. The variation in allele frequency results in differences in metabolic CYP2D6 activity amongst ethnic groups [33].
A translation of CYP2D6 genotype into predicted CYP2D6 phenotype is shown in Table 1. Important changes were published in 2019, where it was internationally agreed to harmonize the CYP2D6*1/*4 interpretation from an Extensive/Normal metabolizer phenotype (CPIC definition until 2017, mostly used in US) into an Intermediate Metabolizer phenotype (definition used by the Dutch Pharmacogenetics Working Group (DPWG), which is mostly used in Europe). The second important change concerns the CYP2D6*10 allele, which was downgraded from AS = 0.5, comparable to other decreased activity alleles such as *9 and *41, to AS = 0.25 [38,39].
A translation of CYP2D6 genotype into predicted CYP2D6 phenotype is shown in Table 1. Important changes were published in 2019, where it was internationally agreed to harmonize the CYP2D6*1/*4 interpretation from an Extensive/Normal metabolizer phenotype (CPIC definition until 2017, mostly used in US) into an Intermediate Metabolizer phenotype (definition used by the Dutch Pharmacogenetics Working Group (DPWG), which is mostly used in Europe). The second important change concerns the CYP2D6*10 allele, which was downgraded from AS = 0.5, comparable to other decreased activity alleles such as *9 and *41, to AS = 0.25 [38][39].
Table 1. Adapted final consensus CYP2D6 genotype to phenotype table. Combining the previous CPIC and DPWG guidelines and adding new pharmacogenetic insights [38]. Abbreviations: CYP2D6: Cytochrome P450 2D6, UM: ultra-rapid metabolizer, NM: normal or extensive metabolizer, IM: intermediate metabolizer, PM: poor metabolizer, CPIC: Clinical Pharmacogenetic Implementation Consortium, DPWG: Dutch Pharmacogenetics Working Group.
. Adapted final consensus CYP2D6 genotype to phenotype table. Combining the previous CPIC and DPWG guidelines and adding new pharmacogenetic insights [38]. Abbreviations: CYP2D6: Cytochrome P450 2D6, UM: ultra-rapid metabolizer, NM: normal or extensive metabolizer, IM: intermediate metabolizer, PM: poor metabolizer, CPIC: Clinical Pharmacogenetic Implementation Consortium, DPWG: Dutch Pharmacogenetics Working Group.
|
Likely Phenotype |
CURRENT CPIC Activity Score Definition |
CURRENT DPWG |
NEW Standardized |
|
CYP2D6 UM |
>2 |
>2.5 |
>2.25 |
|
CYP2D6 NM |
1–2 |
1.5–2.5 |
1.25–2.25 |
|
CYP2D6 IM |
0.5 |
0.5–1.0 |
0.25–1.0 |
|
CYP2D6 PM |
0 |
0 |
0 |
Most recently, Lee et al. suggested a dichotomization into normal and slow metabolizer CYP2D6 groups in an effort to improve and simplify the current system [40]. Nonetheless, these authors also recommend considering the direct measurement of endoxifen plasma concentrations, as CYP2D6 genotype is not solely responsible for systemic endoxifen levels. In line with this recommendation, several authors [41–44][41][42][43][44] as well as the CPIC tamoxifen guideline [17] suggest that individualized dosing (e.g., therapeutic drug monitoring, TDM) might be a better option, instead of using a standard dosage of 20 mg tamoxifen per day. Another option is direct phenotyping, using dextromethorphan as a CYP2D6 phenotyping probe [45–47][45][46][47]. The advantage of this approach is that no genotype to phenotype translation is required, and that dose adjustment can be determined before the start of therapy [47]. Nevertheless, there are many more metabolites with unknown or estrogen-like properties [48]. Therefore, tamoxifen dosing solely based on (predicted) endoxifen blood concentrations might not be the best approach.
References
- Jordan, C. Tamoxifen as the first targeted long-term adjuvant therapy for breast cancer. Endocr. Relat. Cancer 2014, 21, R235–R246.
- Briest, ; Stearns, V. Tamoxifen metabolism and its effect on endocrine treatment of breast cancer. Clin. Adv. Hematol. Oncol. 2009, 7, 185–192.
- Jordan, C. Fourteenth Gaddum Memorial Lecture. A current view of tamoxifen for the treatment and prevention of breast cancer. Br. J. Pharm. 1993, 110, 507–517.
- Osborne, K. Tamoxifen in the treatment of breast cancer. N. Engl. J. Med. 1998, 339, 1609–1618.
- Dean, Tamoxifen therapy and CYP2D6 genotype. In Medical Genetics Summaries [Internet]; Pratt, V.M., Pirmohamed, M., Eds.; National Center for Biotechnology Information (US): Bethesda, MD, USA, 2014.
- Binkhorst, ; Bannink, M.; de Bruijn, P.; Ruit, J.; Droogendijk, H.; van Alphen, R.J.; den Boer, T.D.; Lam, M.H.; Jager, A.; van Gelder, T.; et al. Augmentation of endoxifen exposure in tamoxifen-treated women following SSRI switch. Clin. Pharm. 2016, 55, 249–255.
- Binkhorst, ; Kloth, J.S.L.; de Wit, A.S.; de Bruijn, P.; Lam, M.H.; Chaves, I.; Burger, H.; van Alphen, R.J.; Hamberg, P.; van Schaik, R.H.N.; et al. Circadian variation in tamoxifen pharmacokinetics in mice and breast cancer patients. Breast Cancer Res. Treat. 2015, 152, 119–128.
- Braal, L.; Hussaarts, K.; Seuren, L.; Oomen-de Hoop, E.; de Bruijn, P.; Buck, S.A.J.; Bos, M.; Thijs-Visser, M.F.; Zuetenhorst, H.J.M.; Mathijssen-van Stein, D.; et al. Influence of green tea consumption on endoxifen steady-state concentration in breast cancer patients treated with tamoxifen. Breast Cancer Res. Treat. 2020, 184, 107–113.
- Hussaarts, ; Hurkmans, D.P.; Oomen-de Hoop, E.; van Harten, L.J.; Berghuis, S.; van Alphen, R.J.; Spierings, L.E.A.; van Rossum-Schornagel, Q.C.; Vastbinder, M.B.; van Schaik, R.H.N.; et al. Impact of curcumin (with or without piperine) on the pharmacokinetics of tamoxifen. Cancers 2019, 11, 403, doi:10.3390/cancers11030403.
- Mueller-Schoell, ; Klopp-Schulze, L.; Schroth, W.; Mürdter, T.; Michelet, R.; Brauch, H.; Huisinga, W.; Joerger, M.; Neven, P.; Koolen, S.L.W.; et al. Obesity alters endoxifen plasma levels in young breast cancer patients: a pharmacometric simulation approach. Clin. Pharmacol. Ther. 2020, 108, 661–670, doi:10.1002/cpt.1960.
- Roden, M.; McLeod, H.L.; Relling, M.V.; Williams, M.S.; Mensah, G.A.; Peterson, J.F.; Van Driest, S.L. Pharmacogenomics. Lancet 2019, 394, 521–532.
- Table of Pharmacogenetic Associations. Availabe online: https://www.fda.gov/medical-devices/precision-medicine/table-pharmacogenetic-associations (accessed on 26 October 2020.).
- Henricks, M.; Lunenburg, C.; de Man, F.M.; Meulendijks, D.; Frederix, G.W.J.; Kienhuis, E.; Creemers, G.J.; Baars, A.; Dezentjé, V.O.; Imholz, A.L.T.; et al. DPYD genotype-guided dose individualisation of fluoropyrimidine therapy in patients with cancer: a prospective safety analysis. Lancet Oncol. 2018, 19, 1459–1467.
- Jin, ; Desta, Z.; Stearns, V.; Ward, B.; Ho, H.; Lee, K.H.; Skaar, T.; Storniolo, A.M.; Li, L.; Araba, A.; et al. CYP2D6 genotype, antidepressant use, and tamoxifen metabolism during adjuvant breast cancer treatment. J. Natl. Cancer Inst. 2005, 97, 30–39.
- Goetz, P.; Rae, J.M.; Suman, V.J.; Safgren, S.L.; Ames, M.M.; Visscher, D.W.; Reynolds, C.; Couch, F.J.; Lingle, W.L.; Flockhart, D.A.; et al. Pharmacogenetics of tamoxifen biotransformation is associated with clinical outcomes of efficacy and hot flashes. J. Clin. Oncol. 2005, 23, 9312–9318.
- Rae, M.; Goetz, M.P.; Hayes, D.F.; Ingle, J.N.; Li, L.; Storniolo, A.M.; Stearns, V.; Flockhart, D.A. CYP2D6 genotype and tamoxifen response. Breast Cancer Res. 2005, 7, E6.
- Goetz, P.; Sangkuhl, K.; Guchelaar, H.J.; Schwab, M.; Province, M.; Whirl-Carrillo, M.; Symmans, W.F.; McLeod, H.L.; Ratain, M.J.; Zembutsu, H.; et al. Clinical Pharmacogenetics Implementation Consortium (CPIC) guideline for CYP2D6 and tamoxifen therapy. Clin. Pharmacol. Ther. 2018, 103, 770–777.
- Cardoso, ; Kyriakides, S.; Ohno, S.; Penault-Llorca, F.; Poortmans, P.; Rubio, I.T.; Zackrisson, S.; Senkus, E.; clinicalguidelines@esmo.org, E.G.C.E.a. Early breast cancer: ESMO Clinical Practice Guidelines for diagnosis, treatment and follow-up†. Ann. Oncol. 2019, 30, 1194–1220.
- Regan, M.; Leyland-Jones, B.; Bouzyk, M.; Pagani, O.; Tang, W.; Kammler, R.; Dell'orto, P.; Biasi, M.O.; Thürlimann, B.; Lyng, M.B.; et al. CYP2D6 genotype and tamoxifen response in postmenopausal women with endocrine-responsive breast cancer: the breast international group 1-98 trial. J. Natl. Cancer Inst. 2012, 104, 441–451.
- Rae, M.; Drury, S.; Hayes, D.F.; Stearns, V.; Thibert, J.N.; Haynes, B.P.; Salter, J.; Sestak, I.; Cuzick, J.; Dowsett, M.; et al. CYP2D6 and UGT2B7 genotype and risk of recurrence in tamoxifen-treated breast cancer patients. J. Natl. Cancer Inst. 2012, 104, 452–460.
- Kelly, M.; Pritchard, K.I. CYP2D6 genotype as a marker for benefit of adjuvant tamoxifen in postmenopausal women: lessons learned. J. Natl. Cancer Inst. 2012, 104, 427–428.
- Stanton, , Jr. Re: CYP2D6 genotype and tamoxifen response in postmenopausal women with endocrine-responsive breast cancer: the Breast International Group 1-98 trial. J. Natl. Cancer Inst. 2012, 104, 1265–1266.
- Pharoah, D.; Abraham, J.; Caldas, C. Re: CYP2D6 genotype and tamoxifen response in postmenopausal women with endocrine-responsive breast cancer: the Breast International Group 1-98 trial and Re: CYP2D6 and UGT2B7 genotype and risk of recurrence in tamoxifen-treated breast cancer patients. J. Natl. Cancer Inst. 2012, 104, 1263–1264.
- Nakamura, ; Ratain, M.J.; Cox, N.J.; McLeod, H.L.; Kroetz, D.L.; Flockhart, D.A. Re: CYP2D6 genotype and tamoxifen response in postmenopausal women with endocrine-responsive breast cancer: the Breast International Group 1-98 trial. J. Natl. Cancer Inst. 2012, 104, 1264.
- Province, A.; Goetz, M.P.; Brauch, H.; Flockhart, D.A.; Hebert, J.M.; Whaley, R.; Suman, V.J.; Schroth, W.; Winter, S.; Zembutsu, H.; et al. CYP2D6 genotype and adjuvant tamoxifen: meta-analysis of heterogeneous study populations. Clin. Pharmacol. Ther. 2014, 95, 216–227.
- Jung, A.; Lim, H.S. Association between CYP2D6 genotypes and the clinical outcomes of adjuvant tamoxifen for breast cancer: a meta-analysis. Pharmacogenomics 2014, 15, 49–60.
- Zeng, ; Liu, Y.; Liu, Z.; You, J.; Chen, Z.; Wang, J.; Peng, Q.; Xie, L.; Li, R.; Li, S.; et al. CYP2D6 polymorphisms influence tamoxifen treatment outcomes in breast cancer patients: a meta-analysis. Cancer Chemother. Pharm. 2013, 72, 287–303.
- Lum, W.; Perel, P.; Hingorani, A.D.; Holmes, M.V. CYP2D6 genotype and tamoxifen response for breast cancer: a systematic review and meta-analysis. PLoS ONE 2013, 8, e76648.
- Cronin-Fenton, P.; Damkier, P.; Lash, T.L. Metabolism and transport of tamoxifen in relation to its effectiveness: new perspectives on an ongoing controversy. Future Oncol. 2014, 10, 107–122.
- Cancer Tomorrow. Availabe online: https://gco.iarc.fr/tomorrow/graphic-isotype?type=0&type_sex=0&mode=population&sex=2&populations=900&cancers=20&age_group=value&apc_male=0&apc_female=0&single_unit=500000&print=0 (accessed on 19 September 2020).
- Saladores, H.; Precht, J.C.; Schroth, W.; Brauch, H.; Schwab, M. Impact of metabolizing enzymes on drug response of endocrine therapy in breast cancer. Expert Rev. Mol. Diagn 2013, 13, 349–365.
- Shagufta; Ahmad, Tamoxifen a pioneering drug: An update on the therapeutic potential of tamoxifen derivatives. Eur. J. Med. Chem. 2018, 143, 515–531.
- Bradford, D. CYP2D6 allele frequency in European Caucasians, Asians, Africans and their descendants. Pharmacogenomics 2002, 3, 229–243.
- McKusick, A.; Hamosh, A. Cytochrome P450, Subfamily IID, Polypeptide 6; CYP2D6. Availabe online: https://www.omim.org/entry/124030 (accessed on 29 September 2020).
- Cyp2d6. Availabe online: https://www.pharmvar.org/gene/cyp2d6 (accessed on 29 September 2020).
- Sachse, ; Brockmöller, J.; Bauer, S.; Roots, I. Cytochrome P450 2D6 variants in a Caucasian population: allele frequencies and phenotypic consequences. Am. J. Hum. Genet. 1997, 60, 284–295.
- Hicks, K.; Swen, J.J.; Gaedigk, A. Challenges in CYP2D6 phenotype assignment from genotype data: a critical assessment and call for standardization. Curr. Drug Metab. 2014, 15, 218–232.
- Final Consensus CYP2D6 Genotype to Phenotype Table-March 2019. Availabe online: https://cpicpgx.org/wp-content/uploads/2019/03/Final-Consensus-CYP2D6-genotype-to-phenotype-table_-final_Mar2019.pdf (accessed on 29 September 2020).
- Caudle, E.; Sangkuhl, K.; Whirl-Carrillo, M.; Swen, J.J.; Haidar, C.E.; Klein, T.E.; Gammal, R.S.; Relling, M.V.; Scott, S.A.; Hertz, D.L.; et al. Standardizing CYP2D6 genotype to phenotype translation: consensus recommendations from the Clinical Pharmacogenetics Implementation Consortium and Dutch Pharmacogenetics Working Group. Clin. Transl. Sci. 2020, 13, 116–124.
- Lee, I.; Low, S.K.; Maldonado, R.; Fox, P.; Balakrishnar, B.; Coulter, S.; de Bruijn, P.; Koolen, S.L.W.; Gao, B.; Lynch, J.; et al. Simplified phenotyping of CYP2D6 for tamoxifen treatment using the N-desmethyl-tamoxifen/ endoxifen ratio. Breast 2020, 54, 229–234.
- Binkhorst, ; Mathijssen, R.H.; Jager, A.; van Gelder, T. Individualization of tamoxifen therapy: much more than just CYP2D6 genotyping. Cancer Treat. Rev. 2015, 41, 289–299.
- Binkhorst, ; van Gelder, T.; Mathijssen, R.H. Individualization of tamoxifen treatment for breast carcinoma. Clin. Pharmacol. 2012, 92, 431–433.
- Helland, ; Henne, N.; Bifulco, E.; Naume, B.; Borgen, E.; Kristensen, V.N.; Kvaløy, J.T.; Lash, T.L.; Alnæs, G.I.G.; van Schaik, R.H.; et al. Serum concentrations of active tamoxifen metabolites predict long-term survival in adjuvantly treated breast cancer patients. Breast Cancer Res. 2017, 19, 125.
- de Vries Schultink, H.M.; Huitema, A.D.R.; Beijnen, J.H. Therapeutic Drug Monitoring of endoxifen as an alternative for CYP2D6 genotyping in individualizing tamoxifen therapy. Breast 2018, 42, 38–40.
- de Graan, J.; Teunissen, S.F.; de Vos, F.Y.; Loos, W.J.; van Schaik, R.H.; de Jongh, F.E.; de Vos, A.I.; van Alphen, R.J.; van der Holt, B.; Verweij, J.; et al. Dextromethorphan as a phenotyping test to predict endoxifen exposure in patients on tamoxifen treatment. J. Clin. Oncol. 2011, 29, 3240–3246.
- Opdam, L.; Modak, A.S.; Gelderblom, H.; Guchelaar, H.J. Further characterization of a ¹³C-dextromethorphan breath test for CYP2D6 phenotyping in breast cancer patients on tamoxifen therapy. J. Breath Res. 2015, 9, 026003.
- Gusella, ; Pasini, F.; Corso, B.; Bertolaso, L.; De Rosa, G.; Falci, C.; Modena, Y.; Barile, C.; Da Corte, Z.D.; Fraccon, A.; et al. Predicting steady-state endoxifen plasma concentrations in breast cancer patients by CYP2D6 genotyping or phenotyping. Which approach is more reliable? Pharm. Res. Perspect 2020, 8, e00646.
- Johänning, J.; Kröner, P.; Thomas, M.; Zanger, U.M.; Nörenberg, A.; Eichelbaum, M.; Schwab, M.; Brauch, H.; Schroth, W.; Mürdter, T.E. The formation of estrogen-like tamoxifen metabolites and their influence on enzyme activity and gene expression of ADME genes. Arch. Toxicol. 2018, 92, 1099–1112.
References
- Jordan, V.C. Tamoxifen as the first targeted long-term adjuvant therapy for breast cancer. Endocr. Relat. Cancer 2014, 21, R235–R246.
- Briest, S.; Stearns, V. Tamoxifen metabolism and its effect on endocrine treatment of breast cancer. Clin. Adv. Hematol. Oncol. 2009, 7, 185–192.
- Jordan, V.C. Fourteenth Gaddum Memorial Lecture. A current view of tamoxifen for the treatment and prevention of breast cancer. Br. J. Pharm. 1993, 110, 507–517.
- Osborne, C.K. Tamoxifen in the treatment of breast cancer. N. Engl. J. Med. 1998, 339, 1609–1618.
- Dean, L. Tamoxifen therapy and CYP2D6 genotype. In Medical Genetics Summaries [Internet]; Pratt, V.M., Pirmohamed, M., Eds.; National Center for Biotechnology Information (US): Bethesda, MD, USA, 2014.
- Binkhorst, L.; Bannink, M.; de Bruijn, P.; Ruit, J.; Droogendijk, H.; van Alphen, R.J.; den Boer, T.D.; Lam, M.H.; Jager, A.; van Gelder, T.; et al. Augmentation of endoxifen exposure in tamoxifen-treated women following SSRI switch. Clin. Pharm. 2016, 55, 249–255.
- Binkhorst, L.; Kloth, J.S.L.; de Wit, A.S.; de Bruijn, P.; Lam, M.H.; Chaves, I.; Burger, H.; van Alphen, R.J.; Hamberg, P.; van Schaik, R.H.N.; et al. Circadian variation in tamoxifen pharmacokinetics in mice and breast cancer patients. Breast Cancer Res. Treat. 2015, 152, 119–128.
- Braal, C.L.; Hussaarts, K.; Seuren, L.; Oomen-de Hoop, E.; de Bruijn, P.; Buck, S.A.J.; Bos, M.; Thijs-Visser, M.F.; Zuetenhorst, H.J.M.; Mathijssen-van Stein, D.; et al. Influence of green tea consumption on endoxifen steady-state concentration in breast cancer patients treated with tamoxifen. Breast Cancer Res. Treat. 2020, 184, 107–113.
- Hussaarts, K.; Hurkmans, D.P.; Oomen-de Hoop, E.; van Harten, L.J.; Berghuis, S.; van Alphen, R.J.; Spierings, L.E.A.; van Rossum-Schornagel, Q.C.; Vastbinder, M.B.; van Schaik, R.H.N.; et al. Impact of curcumin (with or without piperine) on the pharmacokinetics of tamoxifen. Cancers 2019, 11, 403.
- Mueller-Schoell, A.; Klopp-Schulze, L.; Schroth, W.; Mürdter, T.; Michelet, R.; Brauch, H.; Huisinga, W.; Joerger, M.; Neven, P.; Koolen, S.L.W.; et al. Obesity alters endoxifen plasma levels in young breast cancer patients: A pharmacometric simulation approach. Clin. Pharmacol. Ther. 2020, 108, 661–670.
- Roden, D.M.; McLeod, H.L.; Relling, M.V.; Williams, M.S.; Mensah, G.A.; Peterson, J.F.; Van Driest, S.L. Pharmacogenomics. Lancet 2019, 394, 521–532.
- FDA. Table of Pharmacogenetic Associations. Available online: (accessed on 26 October 2020).
- Henricks, L.M.; Lunenburg, C.; de Man, F.M.; Meulendijks, D.; Frederix, G.W.J.; Kienhuis, E.; Creemers, G.J.; Baars, A.; Dezentjé, V.O.; Imholz, A.L.T.; et al. DPYD genotype-guided dose individualisation of fluoropyrimidine therapy in patients with cancer: A prospective safety analysis. Lancet Oncol. 2018, 19, 1459–1467.
- Jin, Y.; Desta, Z.; Stearns, V.; Ward, B.; Ho, H.; Lee, K.H.; Skaar, T.; Storniolo, A.M.; Li, L.; Araba, A.; et al. CYP2D6 genotype, antidepressant use, and tamoxifen metabolism during adjuvant breast cancer treatment. J. Natl. Cancer Inst. 2005, 97, 30–39.
- Goetz, M.P.; Rae, J.M.; Suman, V.J.; Safgren, S.L.; Ames, M.M.; Visscher, D.W.; Reynolds, C.; Couch, F.J.; Lingle, W.L.; Flockhart, D.A.; et al. Pharmacogenetics of tamoxifen biotransformation is associated with clinical outcomes of efficacy and hot flashes. J. Clin. Oncol. 2005, 23, 9312–9318.
- Rae, J.M.; Goetz, M.P.; Hayes, D.F.; Ingle, J.N.; Li, L.; Storniolo, A.M.; Stearns, V.; Flockhart, D.A. CYP2D6 genotype and tamoxifen response. Breast Cancer Res. 2005, 7, E6.
- Goetz, M.P.; Sangkuhl, K.; Guchelaar, H.J.; Schwab, M.; Province, M.; Whirl-Carrillo, M.; Symmans, W.F.; McLeod, H.L.; Ratain, M.J.; Zembutsu, H.; et al. Clinical Pharmacogenetics Implementation Consortium (CPIC) guideline for CYP2D6 and tamoxifen therapy. Clin. Pharmacol. Ther. 2018, 103, 770–777.
- Cardoso, F.; Kyriakides, S.; Ohno, S.; Penault-Llorca, F.; Poortmans, P.; Rubio, I.T.; Zackrisson, S.; Senkus, E.; ESMO Guidelines Committee. Early breast cancer: ESMO Clinical Practice Guidelines for diagnosis, treatment and follow-up. Ann. Oncol. 2019, 30, 1194–1220.
- Regan, M.M.; Leyland-Jones, B.; Bouzyk, M.; Pagani, O.; Tang, W.; Kammler, R.; Dell’orto, P.; Biasi, M.O.; Thürlimann, B.; Lyng, M.B.; et al. CYP2D6 genotype and tamoxifen response in postmenopausal women with endocrine-responsive breast cancer: The breast international group 1-98 trial. J. Natl. Cancer Inst. 2012, 104, 441–451.
- Rae, J.M.; Drury, S.; Hayes, D.F.; Stearns, V.; Thibert, J.N.; Haynes, B.P.; Salter, J.; Sestak, I.; Cuzick, J.; Dowsett, M.; et al. CYP2D6 and UGT2B7 genotype and risk of recurrence in tamoxifen-treated breast cancer patients. J. Natl. Cancer Inst. 2012, 104, 452–460.
- Kelly, C.M.; Pritchard, K.I. CYP2D6 genotype as a marker for benefit of adjuvant tamoxifen in postmenopausal women: Lessons learned. J. Natl. Cancer Inst. 2012, 104, 427–428.
- Stanton, V., Jr. Re: CYP2D6 genotype and tamoxifen response in postmenopausal women with endocrine-responsive breast cancer: The Breast International Group 1-98 trial. J. Natl. Cancer Inst. 2012, 104, 1265–1266.
- Pharoah, P.D.; Abraham, J.; Caldas, C. Re: CYP2D6 genotype and tamoxifen response in postmenopausal women with endocrine-responsive breast cancer: The Breast International Group 1-98 trial and Re: CYP2D6 and UGT2B7 genotype and risk of recurrence in tamoxifen-treated breast cancer patients. J. Natl. Cancer Inst. 2012, 104, 1263–1264.
- Nakamura, Y.; Ratain, M.J.; Cox, N.J.; McLeod, H.L.; Kroetz, D.L.; Flockhart, D.A. Re: CYP2D6 genotype and tamoxifen response in postmenopausal women with endocrine-responsive breast cancer: The Breast International Group 1-98 trial. J. Natl. Cancer Inst. 2012, 104, 1264.
- Province, M.A.; Goetz, M.P.; Brauch, H.; Flockhart, D.A.; Hebert, J.M.; Whaley, R.; Suman, V.J.; Schroth, W.; Winter, S.; Zembutsu, H.; et al. CYP2D6 genotype and adjuvant tamoxifen: Meta-analysis of heterogeneous study populations. Clin. Pharmacol. Ther. 2014, 95, 216–227.
- Jung, J.A.; Lim, H.S. Association between CYP2D6 genotypes and the clinical outcomes of adjuvant tamoxifen for breast cancer: A meta-analysis. Pharmacogenomics 2014, 15, 49–60.
- Zeng, Z.; Liu, Y.; Liu, Z.; You, J.; Chen, Z.; Wang, J.; Peng, Q.; Xie, L.; Li, R.; Li, S.; et al. CYP2D6 polymorphisms influence tamoxifen treatment outcomes in breast cancer patients: A meta-analysis. Cancer Chemother. Pharm. 2013, 72, 287–303.
- Lum, D.W.; Perel, P.; Hingorani, A.D.; Holmes, M.V. CYP2D6 genotype and tamoxifen response for breast cancer: A systematic review and meta-analysis. PLoS ONE 2013, 8, e76648.
- Cronin-Fenton, D.P.; Damkier, P.; Lash, T.L. Metabolism and transport of tamoxifen in relation to its effectiveness: New perspectives on an ongoing controversy. Future Oncol. 2014, 10, 107–122.
- WHO. Cancer Tomorrow. Available online: (accessed on 19 September 2020).
- Saladores, P.H.; Precht, J.C.; Schroth, W.; Brauch, H.; Schwab, M. Impact of metabolizing enzymes on drug response of endocrine therapy in breast cancer. Expert Rev. Mol. Diagn. 2013, 13, 349–365.
- Shagufta; Ahmad, I. Tamoxifen a pioneering drug: An update on the therapeutic potential of tamoxifen derivatives. Eur. J. Med. Chem. 2018, 1443, 515–531.
- Bradford, L.D. CYP2D6 allele frequency in European Caucasians, Asians, Africans and their descendants. Pharmacogenomics 2002, 3, 229–243.
- McKusick, V.A.; Hamosh, A. Cytochrome P450, Subfamily IID, Polypeptide 6; CYP2D6. Available online: (accessed on 29 September 2020).
- PharmVar. Cyp2d6. Available online: (accessed on 29 September 2020).
- Sachse, C.; Brockmöller, J.; Bauer, S.; Roots, I. Cytochrome P450 2D6 variants in a Caucasian population: Allele frequencies and phenotypic consequences. Am. J. Hum. Genet. 1997, 60, 284–295.
- Hicks, J.K.; Swen, J.J.; Gaedigk, A. Challenges in CYP2D6 phenotype assignment from genotype data: A critical assessment and call for standardization. Curr. Drug Metab. 2014, 15, 218–232.
- CPIC. Final Consensus CYP2D6 Genotype to Phenotype Table-March. 2019. Available online: (accessed on 29 September 2020).
- Caudle, K.E.; Sangkuhl, K.; Whirl-Carrillo, M.; Swen, J.J.; Haidar, C.E.; Klein, T.E.; Gammal, R.S.; Relling, M.V.; Scott, S.A.; Hertz, D.L.; et al. Standardizing CYP2D6 genotype to phenotype translation: Consensus recommendations from the Clinical Pharmacogenetics Implementation Consortium and Dutch Pharmacogenetics Working Group. Clin. Transl. Sci. 2020, 13, 116–124.
- Lee, C.I.; Low, S.K.; Maldonado, R.; Fox, P.; Balakrishnar, B.; Coulter, S.; de Bruijn, P.; Koolen, S.L.W.; Gao, B.; Lynch, J.; et al. Simplified phenotyping of CYP2D6 for tamoxifen treatment using the N-desmethyl-tamoxifen/ endoxifen ratio. Breast 2020, 54, 229–234.
- Binkhorst, L.; Mathijssen, R.H.; Jager, A.; van Gelder, T. Individualization of tamoxifen therapy: Much more than just CYP2D6 genotyping. Cancer Treat. Rev. 2015, 41, 289–299.
- Binkhorst, L.; van Gelder, T.; Mathijssen, R.H. Individualization of tamoxifen treatment for breast carcinoma. Clin. Pharmacol. 2012, 92, 431–433.
- Helland, T.; Henne, N.; Bifulco, E.; Naume, B.; Borgen, E.; Kristensen, V.N.; Kvaløy, J.T.; Lash, T.L.; Alnæs, G.I.G.; van Schaik, R.H.; et al. Serum concentrations of active tamoxifen metabolites predict long-term survival in adjuvantly treated breast cancer patients. Breast Cancer Res. 2017, 19, 125.
- De Vries Schultink, A.H.M.; Huitema, A.D.R.; Beijnen, J.H. Therapeutic Drug Monitoring of endoxifen as an alternative for CYP2D6 genotyping in individualizing tamoxifen therapy. Breast 2018, 42, 38–40.
- de Graan, A.J.; Teunissen, S.F.; de Vos, F.Y.; Loos, W.J.; van Schaik, R.H.; de Jongh, F.E.; de Vos, A.I.; van Alphen, R.J.; van der Holt, B.; Verweij, J.; et al. Dextromethorphan as a phenotyping test to predict endoxifen exposure in patients on tamoxifen treatment. J. Clin. Oncol. 2011, 29, 3240–3246.
- Opdam, F.L.; Modak, A.S.; Gelderblom, H.; Guchelaar, H.J. Further characterization of a ¹3C-dextromethorphan breath test for CYP2D6 phenotyping in breast cancer patients on tamoxifen therapy. J. Breath Res. 2015, 9, 026003.
- Gusella, M.; Pasini, F.; Corso, B.; Bertolaso, L.; De Rosa, G.; Falci, C.; Modena, Y.; Barile, C.; Da Corte, Z.D.; Fraccon, A.; et al. Predicting steady-state endoxifen plasma concentrations in breast cancer patients by CYP2D6 genotyping or phenotyping. Which approach is more reliable? Pharm. Res. Perspect. 2020, 8, e00646.
- Johänning, J.; Kröner, P.; Thomas, M.; Zanger, U.M.; Nörenberg, A.; Eichelbaum, M.; Schwab, M.; Brauch, H.; Schroth, W.; Mürdter, T.E. The formation of estrogen-like tamoxifen metabolites and their influence on enzyme activity and gene expression of ADME genes. Arch. Toxicol. 2018, 92, 1099–1112.