Non-coding RNA induced by low oxygen partial pressure play a crucial role in cancer progression and therapeutic response.
- Hypoxia
- HIF
- lncRNAs
- miRNAs
- cancer
- proliferation
- cell cycle
1. Introduction
Hypoxia-responsive ncRNAs have been found to play important roles in hypoxia-driven cancer progression modulating the hypoxic gene expression at transcriptional, and post-transcriptional levels, by acting as effectors of HIF or as direct modulators of the HIF-transcriptional cascade [44,45][1][2]. (Figure1)
Profiling techniques and bioinformatics analysis allowed us to unveil more and more hypoxia-regulated non-coding RNA by the presence of the hypoxia response elements (HREs) in their promoter regions [46][3]. Moreover, several studies have described hypoxic induction of non-coding RNAs lacking HREs indicating an indirect regulation often involving epigenetic mechanisms; HIF may control non-coding RNAs expression through histone deacetylase activation, or affecting miRNA maturation machinery [47,48].[4][5]
Using microarray analysis on hypoxia-induced gastric cancer cell lines, Wang et al. identified several hypoxia-responsive lncRNAs in gastric cancer. In particular, they found that an intronic antisense lncRNA named lncRNA-AK058003 was among the most induced lncRNAs upon hypoxia treatment in all examined gastric cancer cell lines [49][6], data confirmed also in breast cancer [50][7]. In addition, recent data demonstrated that HIF-1α can directly regulate circRNAs at the transcriptional level[8][9] [51,52] and that HIF-induced circRNAs may promote cancer growth as demonstrated in bladder [53][10]; however, unlike miRNAs and lncRNAs, the mechanisms of HIF-mediated circRNAs expression have been less investigated and will not be further addressed in this review.
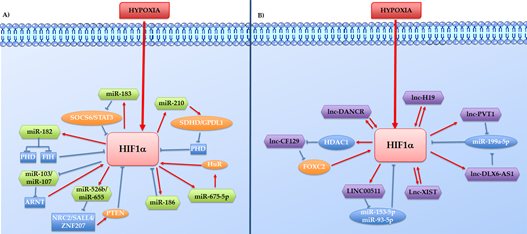
Considering the different mechanisms through which ncRNAs might control tumour growth, these have been divided here into two different groups: 1) the hypoxia-induced ncRNAs that work as HIF effector in promoting cell growth or inhibiting cell death, and 2) the Hypoxia induce ncRNAs such as aHIF-1α, linc-ROR, and lincRNA-p21 which directly or indirectly regulate the HIFs proteins (Figure 2).
Figure 2. Direct or indirect feedback loops between HIF-1a and hypoxia-regulated ncRNAs. The hypoxia-regulated ncRNAs, HIF-1a, and other co-operators intertwine to form reciprocal feedback loops in both positive and negative manners, represented in the figure respectively with red arrows and blue lines. A) Reciprocal feedback loops between HIF-1a and hypoxia-regulated lncRNAs. B) Reciprocal feedback loops between HIF-1a and hypoxia-regulated miRNAs.
2.1. The Hypoxia-Induced ncRNAs as HIF’s Effectors in Controlling Cell Viability
2.1.1. Hypoxia-Induced miRNAs with a Role in Tumour Growth
Hypoxic microenvironment can promote tumour growth in a dual mode: by inducing cell cycle deregulation and by allowing apoptosis escape. Several ncRNAs may act as molecular mediators through which, HIF complex controls these processes. Here we collected the most recent and relevant finding of hypoxia-induced miRNAs (hypoxiamiR), further summarized in Table 1.
Table 1.
List of hypoxia-responsive miRNAs involved in cell proliferation, apoptosis and cell cycle regulation.
miRNAs | Cancer types | Regulation hypoxia-mediated | Targets | Functions | References | ||||||||||||
miR-210 | Schwannoma cells | upregulation | NA | enhances tumour cell proliferation |
[54] |
[11] |
|||||||||||
neuroblastoma cells | upregulation | Bcl-2 | induces apoptosis |
[55] |
[12] |
||||||||||||
Breast and melanoma cancer cells, | upregulation | Max’s Next Tango (MNT) | inhibits hypoxia-induced cell cycle arrest |
[56] |
[13] |
||||||||||||
Glioma stem cells | upregulation | MNT-Max complex | inhibits hypoxia-induced cell cycle arrest |
[57] |
[14] |
||||||||||||
Epithelial ovarian cancer | upregulation | PTPN1 | promotes cell proliferation and inhibits apoptosis |
[58] |
[15] |
||||||||||||
Epatoma cells | upregulation | AIFM3 | inhibits hypoxia-induced cell cycle arrest |
[59] |
[16] |
| |||||||||||
Glioma cells | upregulation | SIN3A | inhibits proliferation and promotes apoptosis |
[60] |
[17] |
||||||||||||
triple-negative breast cancer | upregulation | p53 | promotes cell proliferation |
[61] |
[18] |
||||||||||||
miR-210-3p | bladder cancer | upregulation | NA | induces apoptosis |
[62] |
[19] |
|||||||||||
miR-145 | breast cancer | upregulation | TGFb2, HuR | promotes proliferation |
[63] |
[20] |
|||||||||||
miR-191 | non-small cell lung cancer cells | upregulation | NF1A | promotes proliferation |
[64] |
[21] |
|||||||||||
gastric cancer | upregulation | MDR1/P-gp, LRP and Bcl-2 pathways | promotes proliferation |
[65] |
[22] |
||||||||||||
miR-27a | gastric cancer | upregulation | PTEN | promotes proliferation |
[66] |
[23] |
|||||||||||
miR-382 | breast cancer | upregulation | PDCD4 | inhibits apoptosis |
[67] |
[24] |
|||||||||||
miR-424 | colorectal cancer | upregulation | DAPK, KLF4 | promotes hyperproliferation and decreases apoptosis |
[68] |
[25] |
|||||||||||
miR-103/107 | pancreatic cancer cells | upregulation | NA | promotes proliferation and inhibits apoptosis |
[69] |
[26] |
|||||||||||
miR-21 | cervical cancer cells | upregulation | PTEN/AKT pathway | promotes cell growth |
[70] |
[27] |
|||||||||||
gastric cancer | upregulation | RASSF8 | promotes cell growth |
[71] |
[28] |
||||||||||||
miR-224 | bladder cancer cells | downregulation | FGFR3 | upregulates proliferation |
[72] |
[29] |
|||||||||||
miR-100 | pancreatic cancer cells | downregulation | Vimentin | inhibits proliferation |
[73] |
[30] |
|||||||||||
miR-548an | acute myeloid leukaemia cells | downregulation | p21, STAT3 | inhibits cell growth |
[74] |
[31] |
|||||||||||
miR-101 | glioblastoma | upregulation | NA | promotes cell proliferation |
[75,76] | ||||||||||||
miR-675 | colorectal cancer cells | upregulation | b-catenin localization | regulates cell cycle |
[77] [78] | ||||||||||||
non-small cell lung cancer | upregulation | p53 | promotes cell proliferation |
[79] |
[36] |
||||||||||||
gastric cancer | upregulation | Caspase 3 | inhibits apoptosis |
[80] |
[37] |
||||||||||||
miR-421 | hepatocellular carcinoma | downregulation | VASP | promotes cell growth |
[81] |
[38] |
|||||||||||
miR-204 | hepatocellular carcinoma | downregulation | TWIST1 | induces tumour cell proliferation |
[82,83] | ||||||||||||
miR-33a | hepatocellular carcinoma | downregulation | NA | upregulates tumour cell proliferation |
[82,83] |
The most studied hypoxiamiR is the miR-210, it is regulated by HIF in various cell types through the direct binding of the transcription factor to the HREs on its promoter. It was found to represses genes expressed under normoxia, and required to promote tumour growth [84][41]. The upregulation of miR-210 in solid tumours was associated with bad prognosis, indicating that the target genes affected by miR-210 have a functional impact on tumour malignancy and drug resistance [85–87][42][43]. Several studies, to date, have demonstrated its involvement in various kinds of tumours affecting a large number of cellular functions, including mitochondrial metabolism, angiogenesis, DNA repair, and cell survival. Wang et al. showed that, in schwannoma cells, hypoxia-induced miR-210 promotes autophagy activation, tumour cell proliferation and angiogenesis, while inhibits apoptosis; intriguingly, in this study, the authors noted that miR-210 promoter region, containing the HREs, was hypermethylated in normoxia, while demethylated in hypoxia thus suggesting a double control on miR-210 expression [54][11]. Concerning the molecular mediators, firstly, the Grandori group identified in breast and melanoma cancer cells that hypoxia-induced miR-210 targets the Max’s Next Tango (MNT) mRNA, a key transcriptional repressor of the MYC-MAX network. MNT downregulation allows c-MYC to push cells through the cell cycle [56][13]. Similar results were obtained by Yang and colleagues in glioma stem cells, in which they observed that hypoxia upregulated miR-210 avoided G0/G1 cell cycle arrest via MNT-Max complex-dependent transcription repression [57][14]. In an in vitro model of ovarian cancer, Li et al. demonstrated that hypoxia upregulates miR-210 thus promoting tumour cell proliferation and cell clone generation via targeting PTPN1 (tyrosine-protein phosphatase non-receptor type 1) and inhibiting apoptosis [58][15]. It is notable that several studies highlighted the predicted miR-210 seed sites in apoptosis-related mRNA transcripts such as AIFM3 (Apoptosis-Inducing Factor Mitochondrion-associated 3), CASP8AP2 (Caspase-8-Associated Protein-2) and SIN3A (a transcription repressor that forms a complex with histone deacetylase 1) [59,60,88,89][16][17][44][45].
Recently, an interesting manuscript of Du et al. demonstrated that hypoxia-induced miR-210-3p controls cell proliferation by promoting Warburg effects and, concomitantly by inhibiting p53 activity in triple-negative breast cancer [61][18].
Taken together, these studies support a model in which, through the regulation of a single miRNA, HIF can simultaneously target multiple factors with a key role in the apoptotic process, ultimately promoting uncontrolled cell proliferation and carcinogenesis.
Another highly expressed hypoxiamir, regulated by both HIF-1 a and HIF-2a, is miR-21. It is considered to act as oncomirs by targeting many tumour suppressor genes involved in cell proliferation, apoptosis, and invasion in several types of cancer [69,70][26][27]. The pro-oncogenic role of miR-21 was in deep studied in the last years since to propose anti-miR-21 as a strategy to fight tumour growth [90][46]; here is interesting to note that hypoxia-induced miR-21 shows an elevated level in hypoxic exosomes. Li et al., demonstrated that tumour-derived exosomes enriched in miR-21 are internalized by normoxic cells driving recipient cells toward a pro-metastatic phenotype [91][47].
One of the miRNAs, which expression increases in the hypoxic tumour, is the miR-675. It was found up-regulated by HIF in several tumours including Glioblastoma[32][33] [75,76] and colorectal cancer [77][34]. Recently it was described its role in controlling cell cycle by regulating Glycogen Synthase Kinase 3b (GSK-3b) activity and allowing b-catenin nuclear localization [78][35]. MiR-675 was found up-regulated in hypoxic non-small cell lung cancer where in addition to promoting cell proliferation, it acts on apoptosis by directly inhibiting the expression of p53 [79][36]; also in this case, through the induction of a single miRNA, HIF promotes tumour growth by acting simultaneously on several molecular pathways.
In a study conducted by Zhao et al., miR-191 was found to be upregulated by HIF-1a. They showed that miR-191 promoted the proliferation and migration of non-small cell lung cancer cells (NSCLC) by targeting of NF1A (Nuclear factor 1 A-type) under chronic hypoxic conditions [64][21]. In breast cancer, Nagpal and colleagues showed that miR-191 is upregulated by both HIF-1a and HIF-2a and its overexpression is responsible for cancer aggressiveness by promoting cell proliferation, and survival under hypoxia; moreover, they demonstrated that miR-191 promotes TGFb expression thus revealing a molecular link between HIF and TGFb signalling pathways, both pivotal in the regulation of breast cancer metastasis [63][20].
He et al. highlighted the role of HIF-1α in transcriptional upregulation of the miR-224 in gastric cancer cells. Through target gene validation, these authors revealed that miR-224 directly targets RASFF8 (Ras association domain family member 8), stimulating p65 nuclear translocation and NF-kB transcriptional activity to confer gastric cancer cells with more aggressive phenotype [71][28]. Their results suggest that hypoxia-inducible miR-224 promotes gastric cancer cell growth by downregulating RASSF8 and acts as an oncogene, implying that inhibition of miR-224 may have potential as a therapeutic target for patients with hypoxic gastric tumours [71][28].
Several studies conducted on gastric cancer cells using different approaches (i.e., ChIP assay, luciferase assay, as well as qRT-PCR) revealed the direct relationship between HIF-1a and different miRNAs. Ge et al. observed that miR-421, up-regulated by HIF-1α, can promote tumour behaviour in gastric cancer by targeting the caspase-3 thus inhibiting apoptosis [80][37]. Zhao and colleagues revealed that HIF-1α can directly bind the miR-27a promoter increasing the activity of some antiapoptotic pathways (i.e., the MDR1/P-gp, Bcl-2, LRP) and promoting multidrug resistance [65][22].
It is notable that while several miRNAs are directly induced by HIF, a great number of non-coding RNAs were found to be down-regulated in hypoxic conditions.
Liu et al. showed that in hepatocellular carcinoma miR-204 expression is inhibited by HIF-1a. These authors proved that hypoxia-induced down-regulation of miR-204 promotes malignant transformation through the up-regulation of Vasodilator-stimulated phosphoprotein (VASP); this is a regulator of cytoskeletal actin and cell migration, and its expression correlates with aggressive phenotype and metastasis both in vitro and in vivo [81][38]. MiR-33a and miR-199a-5p are found to be reduced under the regulation of HIF-1a in HCC [82,83][39][40]. Li et al. proved that mirR-199a-5p inhibition by HIF-1a regulates Warburg effect and induce tumour cell proliferation in hepatocellular carcinoma [83][40]. Also, miR-548an, a tumour suppressor miRNA, is down-regulated by HIF-1a in pancreatic cancer cells, and it is involved in increasing vimentin level and facilitating the pancreatic tumorigenesis [73][30].
Intriguingly, several studies have reported hypoxamirs working as tumour suppressors e.g., the miR-145 or the miR-215 [62,92][19][48], thus suggesting the existence of an intricate network of interactors not yet fully revealed. To solve this, some aspects that have been neglected so far must be taken into account: i) here we reviewed only a fraction of HIF-induced miRNA but several other miRNAs are induced under hypoxic condition in a HIF-independent manner, ii) a single miRNA has numerous targets in the same cell, and iii) hypoxia-induced lncRNAs can sequester multiple miRNAs preventing them from reaching their targets.
Reference (Editors will rearrange the references after the entry is submitted)
- Najafi, ; Farhood, B.; Mortezaee, K.; Kharazinejad, E.; Majidpoor, J.; Ahadi, R. Hypoxia in solid tumors: A key promoter of cancer stem cell (CSC) resistance. J. Cancer Res. Clin. Oncol. 2020, 146, 19–31, doi:10.1007/s00432-019-03080-1.
- Kim, -C.; Hwang, S.-H.; Kim, N.-Y.; Lee, H.-S.; Ji, S.; Yang, Y.; Kim, Y. Hypoxia promotes acquisition of aggressive phenotypes in human malignant mesothelioma. BMC Cancer 2018, 18, 1–11, doi:10.1186/s12885-018-4720-z.
- Xiang, ; Semenza, G.L. Hypoxia-inducible factors promote breast cancer stem cell specification and maintenance in response to hypoxia or cytotoxic chemotherapy. Adv. Cancer Res. 2019, 141, 175–212, doi:10.1016/bs.acr.2018.11.001.
- Wu, ; Potluri, N.; Lu, J.; Kim, Y.; Rastinejad, F. Structural integration in hypoxia-inducible factors. Nat. Cell Biol. 2015, 524, 303–308, doi:10.1038/nature14883.
- Smythies, J.A.; Sun, M.; Masson, N.; Salama, R.; Simpson, P.D.; Murray, E.; Neumann, V.; Cockman, M.E.; Choudhry, H.; Ratcliffe, P.J.; et al. Inherent DNA ‐binding specificities of the HIF ‐1α and HIF ‐2α transcription factors in EMBO Rep. 2019, 20, doi:10.15252/embr.201846401.
- Maynard, A.; Evans, A.J.; Hosomi, T.; Hara, S.; Jewett, M.A.S.; Ohh, M. Human HIF‐3α4 is a dominant‐negative regulator of HIF‐1 and is down‐regulated in renal cell carcinoma. FASEB J. 2005, 19, 1396–1406, doi:10.1096/fj.05-3788com.
- Yamashita, ; Ohneda, O.; Nagano, M.; Iemitsu, M.; Makino, Y.; Tanaka, H.; Miyauchi, T.; Goto, K.; Ohneda, K.; Fujii-Kuriyama, Y.; et al. Abnormal Heart Development and Lung Remodeling in Mice Lacking the Hypoxia-Inducible Factor-Related Basic Helix-Loop-Helix PAS Protein NEPAS. Mol. Cell. Biol. 2007, 28, 1285–1297, doi:10.1128/mcb.01332-07.
- Kobayashi, ; Yamashita, T.; Ohneda, K.; Nagano, M.; Kimura, K.; Nakai, H.; Poellinger, L.; Ohneda, O. Hypoxia-inducible factor-3α promotes angiogenic activity of pulmonary endothelial cells by repressing the expression of the VE-cadherin gene. Genes Cells 2015, 20, 224–241, doi:10.1111/gtc.12215.
- Tolonen, -P.; Heikkilä, M.; Malinen, M.; Lee, H.-M.; Palvimo, J.J.; Wei, G.-H.; Myllyharju, J. A long hypoxia-inducible factor 3 isoform 2 is a transcription activator that regulates erythropoietin. Cell. Mol. Life Sci. 2020, 77, 3627–3642, doi:10.1007/s00018-019-03387-9.
- Depping, ; Jelkmann, W.; Kosyna, F.K. Nuclear-cytoplasmatic shuttling of proteins in control of cellular oxygen sensing. J. Mol. Med. 2015, 93, 599–608, doi:10.1007/s00109-015-1276-0.
- Semenza, L. Hypoxia‐inducible factors: Coupling glucose metabolism and redox regulation with induction of the breast cancer stem cell phenotype. EMBO J. 2017, 36, 252–259, doi:10.15252/embj.201695204.
- Lv, ; Li, J.; Zhang, C.; Hu, T.; Li, S.; He, S.; Yan, H.; Tan, Y.; Lei, M.; Wen, M.; et al. The role of hypoxia-inducible factors in tumor angiogenesis and cell metabolism. Genes Dis. 2017, 4, 19–24, doi:10.1016/j.gendis.2016.11.003.
- Hubbi, E.; Semenza, G.L. Regulation of cell proliferation by hypoxia-inducible factors. Am. J. Physiol.-Cell Physiol. 2015, 309, C775–C782, doi:10.1152/ajpcell.00279.2015.
- Vaddi, R.; Piao, L.; Khan, S.A.; Wang, N.; Prabhakar, N.R.; Nanduri, J. Hypoxia induced hERG trafficking defect linked to cell cycle arrest in SH-SY5Y cells. PLoS ONE 2019, 14, e0215905, doi:10.1371/journal.pone.0215905.
- Ha, ; Kim, V.N. Regulation of microRNA biogenesis. Nat. Rev. Mol. Cell Biol. 2014, 15, 509–524, doi:10.1038/nrm3838.
- Han, ; Lee, Y.; Yeom, K.-H.; Kim, Y.-K.; Jin, H.; Kim, V.N. The Drosha-DGCR8 complex in primary microRNA processing. Genes Dev. 2004, 18, 3016–3027, doi:10.1101/gad.1262504.
- Berg, V.D.; Mols, J.; Han, J. RISC-target interaction: Cleavage and translational suppression. Biochim. Biophys. Acta BBA Bioenerg. 2008, 1779, 668–677, doi:10.1016/j.bbagrm.2008.07.005.
- Place, F.; Li, L.-C.; Pookot, D.; Noonan, E.J.; Dahiya, R. MicroRNA-373 induces expression of genes with complementary promoter sequences. Proc. Natl. Acad. Sci. USA 2008, 105, 1608–1613, doi:10.1073/pnas.0707594105.
- Orang, V.; Safaralizadeh, R.; Kazemzadeh-Bavili, M. Mechanisms of miRNA-Mediated Gene Regulation from Common Downregulation to mRNA-Specific Upregulation. Int. J. Genom. 2014, 2014, 1–15, doi:10.1155/2014/970607.
- De Sousa, C.; Gjorgjieva, M.; Dolicka, D.; Sobolewski, C.; Foti, M. Deciphering miRNAs’ Action through miRNA Editing. Int. J. Mol. Sci. 2019, 20, 6249, doi:10.3390/ijms20246249.
- Schmitz, U.; Grote, P.; Herrmann, B.G. Mechanisms of long noncoding RNA function in development and disease. Cell. Mol. Life Sci. 2016, 73, 2491–2509, doi:10.1007/s00018-016-2174-5.
- Ma, ; Bajic, V.B.; Zhang, Z. On the classification of long non-coding RNAs. RNA Biol. 2013, 10, 924–933, doi:10.4161/rna.24604.
- Quinn, J.; Chang, H.Y. Unique features of long non-coding RNA biogenesis and function. Nat. Rev. Genet. 2016, 17, 47–62, doi:10.1038/nrg.2015.10.
- Krchňáková, ; Thakur, P.K.; Krausová, M.; Bieberstein, N.; Haberman, N.; Müller-McNicoll, M.; Staněk, D. Splicing of long non-coding RNAs primarily depends on polypyrimidine tract and 5′ splice-site sequences due to weak interactions with SR proteins. Nucleic Acids Res. 2019, 47, 911–928, doi:10.1093/nar/gky1147.
- Jiang, ; Agrawal, D.K.; Boosani, C.S. Non-coding RNAs as Epigenetic Gene Regulators in Cardiovascular Diseases. Adv. Exp. Med. Biol. 2020, 1229, 133–148, doi:10.1007/978-981-15-1671-9_7.
- Zhang, ; Wang, W.; Zhu, W.; Dong, J.; Cheng, Y.; Yin, Z.; Shen, F. Mechanisms and Functions of Long Non-Coding RNAs at Multiple Regulatory Levels. Int. J. Mol. Sci. 2019, 20, 5573, doi:10.3390/ijms20225573.
- Li, ; Guo, G.; Lu, M.; Chai, W.; Li, Y.; Tong, X.; Li, J.; Jia, X.; Liu, W.; Qi, D.; et al. Long Noncoding RNA Lnc-MxA Inhibits Beta Interferon Transcription by Forming RNA-DNA Triplexes at Its Promoter. J. Virol. 2019, 93, doi:10.1128/jvi.00786-19.
- Romero-Barrios, ; Legascue, M.F.; Benhamed, M.; Ariel, F.; Crespi, M. Splicing regulation by long noncoding RNAs. Nucleic Acids Res. 2018, 46, 2169–2184, doi:10.1093/nar/gky095.
- Jonas, ; Calin, G.A.; Pichler, M. RNA-Binding Proteins as Important Regulators of Long Non-Coding RNAs in Cancer. Int. J. Mol. Sci. 2020, 21, 2969, doi:10.3390/ijms21082969.
- Zhang, ; Liu, L.; Li, J.; Le, T.D. LncmiRSRN: Identification and analysis of long non-coding RNA related miRNA sponge regulatory network in human cancer. Bioinformatics 2018, 34, 4232–4240, doi:10.1093/bioinformatics/bty525.
- Salzman, Circular RNA Expression: Its Potential Regulation and Function. Trends Genet. 2016, 32, 309–316, doi:10.1016/j.tig.2016.03.002.
- Vo, N.; Cieslik, M.; Zhang, Y.; Shukla, S.; Xiao, L.; Zhang, Y.; Wu, Y.-M.; Dhanasekaran, S.M.; Engelke, C.G.; Cao, X.; et al. The Landscape of Circular RNA in Cancer. Cell 2019, 176, 869–881, doi:10.1016/j.cell.2018.12.021.
- Jeck, R.; Sorrentino, J.A.; Wang, K.; Slevin, M.K.; Burd, C.E.; Liu, J.; Marzluff, W.F.; Sharpless, N.E. Circular RNAs are abundant, conserved, and associated with ALU repeats. RNA 2012, 19, 141–157, doi:10.1261/rna.035667.112.
- Abe, ; Matsumoto, K.; Nishihara, M.; Nakano, Y.; Shibata, A.; Maruyama, H.; Shuto, S.; Matsuda, A.; Yoshida, M.; Ito, Y.; et al. Rolling Circle Translation of Circular RNA in Living Human Cells. Sci. Rep. 2015, 5, 16435, doi:10.1038/srep16435.
- Legnini, ; Di Timoteo, G.; Rossi, F.; Morlando, M.; Briganti, F.; Sthandier, O.; Fatica, A.; Santini, T.; Andronache, A.; Wade, M.; et al. Circ-ZNF609 Is a Circular RNA that Can Be Translated and Functions in Myogenesis. Mol. Cell 2017, 66, 22–37, doi:10.1016/j.molcel.2017.02.017.
- Lei, ; Zheng, G.; Ning, Q.; Zheng, J.; Dong, D. Translation and functional roles of circular RNAs in human cancer. Mol. Cancer 2020, 19, 1–9, doi:10.1186/s12943-020-1135-7.
- Li, ; Huang, C.; Bao, C.; Chen, L.; Lin, M.; Wang, X.; Zhong, G.; Yu, B.; Hu, W.; Dai, L.; et al. Exon-intron circular RNAs regulate transcription in the nucleus. Nat. Struct. Mol. Biol. 2015, 22, 256–264, doi:10.1038/nsmb.2959.
- Zhang, ; Zhang, X.-O.; Chen, T.; Xiang, J.-F.; Yin, Q.-F.; Xing, Y.-H.; Zhu, S.; Yang, L.; Chen, L.-L. Circular Intronic Long Noncoding RNAs. Mol. Cell 2013, 51, 792–806, doi:10.1016/j.molcel.2013.08.017.
- Lu, ; Filonov, G.S.; Noto, J.J.; Schmidt, C.A.; Hatkevich, T.L.; Wen, Y.; Jaffrey, S.R.; Matera, A.G. Metazoan tRNA introns generate stable circular RNAs in vivo. RNA 2015, 21, 1554–1565, doi:10.1261/rna.052944.115.
- Hansen, B.; Jensen, T.I.; Clausen, B.H.; Bramsen, J.B.; Finsen, B.; Damgaard, C.K.; Kjems, J. Natural RNA circles function as efficient microRNA sponges. Nat. Cell Biol. 2013, 495, 384–388, doi:10.1038/nature11993.
- Lasda, ; Parker, R. Circular RNAs: Diversity of form and function. RNA 2014, 20, 1829–1842, doi:10.1261/rna.047126.114.
- Xu, ; Xie, F.; Tang, X.; Wang, T.; Wang, S. Insights into the role of circular RNA in macrophage activation and fibrosis disease. Pharmacol. Res. 2020, 156, 104777, doi:10.1016/j.phrs.2020.104777.
- Huang, ; Zheng, H.; Wu, Z.; Chen, M.; Huang, Y. Circular RNA-protein interactions: Functions, mechanisms, and identification. Theranostics 2020, 10, 3503–3517, doi:10.7150/thno.42174.
- Camps, ; Saini, H.K.; Mole, D.R.; Choudhry, H.; Reczko, M.; Guerra-Assunção, J.A.; Tian, Y.-M.; Buffa, F.M.; Harris, A.L.; Hatzigeorgiou, A.G.; et al. Integrated analysis of microRNA and mRNA expression and association with HIF binding reveals the complexity of microRNA expression regulation under hypoxia. Mol. Cancer 2014, 13, 28, doi:10.1186/1476-4598-13-28.
- Nallamshetty, ; Chan, S.Y.; Loscalzo, J. Hypoxia: A master regulator of microRNA biogenesis and activity. Free Radic. Biol. Med. 2013, 64, 20–30, doi:10.1016/j.freeradbiomed.2013.05.022.
- Slemc, ; Kunej, T. Transcription factor HIF1A: Downstream targets, associated pathways, polymorphic hypoxia response element (HRE) sites, and initiative for standardization of reporting in scientific literature. Tumor Biol. 2016, 37, 14851–14861, doi:10.1007/s13277-016-5331-4.
- Choudhry, ; Schödel, J.; Oikonomopoulos, S.; Camps, C.; Grampp, S.; Harris, A.L.; Ratcliffe, P.J.; Ragoussis, J.; Mole, D.R. Extensive regulation of the non‐coding transcriptome by hypoxia: Role of HIF in releasing paused RNA pol2. EMBO Rep. 2014, 15, 70–76, doi:10.1002/embr.201337642.
- Lai, -H.; Li, J.-N.; Wang, M.-Y.; Huang, H.-Y.; Croce, C.M.; Sun, H.-L.; Lyu, Y.-J.; Kang, J.-W.; Chiu, C.-F.; Hung, M.-C.; et al. HIF-1α promotes autophagic proteolysis of Dicer and enhances tumor metastasis. J. Clin. Investig. 2017, 128, 625–643, doi:10.1172/jci89212.
- Wang, ; Liu, X.; Zhang, H.; Sun, L.; Zhou, Y.; Jin, H.; Zhang, H.; Zhang, H.; Liu, J.; Guo, H.; et al. Hypoxia-inducible lncRNA-AK058003 promotes gastric cancer metastasis by targeting γ-synuclein. Neoplasia 2014, 16, 1094–1106, doi:10.1016/j.neo.2014.10.008.
- He, ; Wang, P. Unregulated long non-coding RNA-AK058003 promotes the proliferation, invasion and metastasis of breast cancer by regulating the expression levels of the γ-synuclein gene. Exp. Ther. Med. 2015, 9, 1727–1732, doi:10.3892/etm.2015.2323.
- Cheng, ; Qiu, J.; Wang, S.; Yang, Y.; Guo, M.; Wang, D.; Luo, Q.; Xu, L. Comprehensive circular RNA profiling identifies CircFAM120A as a new biomarker of hypoxic lung adenocarcinoma. Ann. Transl. Med. 2019, 7, 442, doi:10.21037/atm.2019.08.79.
- Di Liddo, ; Machado, C.D.O.F.; Fischer, S.; Ebersberger, S.; Heumüller, A.W.; E Weigand, J.; Müller-McNicoll, M.; Zarnack, K. A combined computational pipeline to detect circular RNAs in human cancer cells under hypoxic stress. J. Mol. Cell Biol. 2019, 11, 829–844, doi:10.1093/jmcb/mjz094.
- Wei, ; Zhang, Y.; Meng, Q.; Cui, L.; Xu, C. Hypoxia-induced circular RNA has_circRNA_403658 promotes bladder cancer cell growth through activation of LDHA. Am. J. Transl Res. 2019, 11, 6838–6849.
- Wang, ; Deng, M.; Liu, Z.; Wu, S. Hypoxia-induced miR-210 promoter demethylation enhances proliferation, autophagy and angiogenesis of schwannoma cells. Oncol. Rep. 2017, 37, 3010–3018, doi:10.3892/or.2017.5511.
- Chio, -C.; Lin, J.-W.; Cheng, H.-A.; Chiu, W.-T.; Wang, Y.-H.; Wang, J.-J.; Hsing, C.-H.; Chen, R.-M. MicroRNA-210 targets antiapoptotic Bcl-2 expression and mediates hypoxia-induced apoptosis of neuroblastoma cells. Arch. Toxicol. 2012, 87, 459–468, doi:10.1007/s00204-012-0965-5.
- Zhang, ; Sun, H.; Dai, H.; Walsh, R.M.; Imakura, M.; Schelter, J.; Burchard, J.; Dai, X.; Chang, A.N.; Diaz, R.L.; et al. MicroRNA miR-210 modulates cellular response to hypoxia through the MYC antagonist MNT. Cell Cycle 2009, 8, 2756–2768, doi:10.4161/cc.8.17.9387.
- Yang, ; Wei, J.; Guo, T.; Shen, Y.; Liu, F. Knockdown of miR-210 decreases hypoxic glioma stem cells stemness and radioresistance. Exp. Cell Res. 2014, 326, 22–35, doi:10.1016/j.yexcr.2014.05.022.
- Li, ; Huang, K.; You, Y.; Fu, X.; Hu, L.; Song, L.; Meng, Y. Hypoxia-induced miR-210 in epithelial ovarian cancer enhances cancer cell viability via promoting proliferation and inhibiting apoptosis. Int. J. Oncol. 2014, 44, 2111–2120, doi:10.3892/ijo.2014.2368.
- Yang, ; Sun, T.; Cao, J.; Liu, F.; Tian, Y.; Zhu, W. Downregulation of miR-210 expression inhibits proliferation, induces apoptosis and enhances radiosensitivity in hypoxic human hepatoma cells in vitro. Exp. Cell Res. 2012, 318, 944–954, doi:10.1016/j.yexcr.2012.02.010.
- Xue, -X.; Shang, C.; Hong, Y.; Guo, Y.; Liu, Y.-H. MiR-210 Up-Regulation Inhibits Proliferation and Induces Apoptosis in Glioma Cells by Targeting SIN3A. Med. Sci. Monit. 2014, 20, 2571–2577, doi:10.12659/msm.892994.
- Du, ; Wei, N.; Ma, R.; Jiang, S.; Song, D. A miR-210-3p regulon that controls the Warburg effect by modulating HIF-1α and p53 activity in triple-negative breast cancer. Cell Death Dis. 2020, 11, 1–12, doi:10.1038/s41419-020-02952-6.
- Blick, ; Ramachandran, A.V.; McCormick, R.; Wigfield, S.; Cranston, D.; Catto, J.W.F.; Harris, A.L. Identification of a hypoxia-regulated miRNA signature in bladder cancer and a role for miR-145 in hypoxia-dependent apoptosis. Br. J. Cancer 2015, 113, 634–644, doi:10.1038/bjc.2015.203.
- Nagpal, ; Ahmad, H.M.; Chameettachal, S.; Sundar, D.; Ghosh, S.; Kulshreshtha, R. HIF-inducible miR-191 promotes migration in breast cancer through complex regulation of TGFβ-signaling in hypoxic microenvironment. Sci. Rep. 2015, 5, 9650, doi:10.1038/srep09650.
- Zhao, ; Qiao, C.-R.; Ding, Z.; Sheng, Y.-L.; Li, X.-N.; Yang, Y.; Zhu, D.-Y.; Zhang, C.-Y.; Liu, D.-L.; Wu, K.; et al. A novel pathway in NSCLC cells: miR-191, targeting NFIA, is induced by chronic hypoxia, and promotes cell proliferation and migration. Mol. Med. Rep. 2017, 15, 1319–1325, doi:10.3892/mmr.2017.6100.
- Zhao, ; Li, Y.; Tan, B.-B.; Fan, L.-Q.; Yang, P.-G.; Tian, Y. HIF-1α Induces Multidrug Resistance in Gastric Cancer Cells by Inducing MiR-27a. PLoS ONE 2015, 10, e0132746, doi:10.1371/journal.pone.0132746.
- Seok, -K.; Lee, S.H.; Kim, M.J.; Lee, Y.-M. MicroRNA-382 induced by HIF-1α is an angiogenic miR targeting the tumor suppressor phosphatase and tensin homolog. Nucleic Acids Res. 2014, 42, 8062–8072, doi:10.1093/nar/gku515.
- Zhang, ; Shi, Z.; Li, M.; Mi, J. Hypoxia-induced miR-424 decreases tumor sensitivity to chemotherapy by inhibiting apoptosis. Cell Death Dis. 2014, 5, e1301, doi:10.1038/cddis.2014.240.
- Chen, -Y.; Lin, Y.-M.; Chung, H.-C.; Lang, Y.-D.; Lin, C.-J.; Huang, J.; Wang, W.-C.; Lin, F.-M.; Chen, Z.; Huang, H.-D.; et al. miR-103/107 Promote Metastasis of Colorectal Cancer by Targeting the Metastasis Suppressors DAPK and KLF4. Cancer Res. 2012, 72, 3631–3641, doi:10.1158/0008-5472.can-12-0667.
- Mace, A.; Collins, A.L.; Wojcik, S.E.; Croce, C.M.; Lesinski, G.B.; Bloomston, M. Hypoxia induces the overexpression of microRNA-21 in pancreatic cancer cells. J. Surg. Res. 2013, 184, 855–860, doi:10.1016/j.jss.2013.04.061.
- Song, ; Liu, S.; Zhang, L.; Yao, H.; Gao, F.; Xu, D.; Lili, S. MiR-21 modulates radiosensitivity of cervical cancer through inhibiting autophagy via the PTEN/Akt/HIF-1α feedback loop and the Akt-mTOR signaling pathway. Tumor Biol. 2016, 37, 12161–12168, doi:10.1007/s13277-016-5073-3.
- He, ; Wang, L.; Zhang, J.; Xu, H. Hypoxia-inducible microRNA-224 promotes the cell growth, migration and invasion by directly targeting RASSF8 in gastric cancer. Mol. Cancer 2017, 16, 1–14, doi:10.1186/s12943-017-0603-1.
- Blick, ; Ramachandran, A.; Wigfield, S.; McCormick, R.; Jubb, A.M.; Buffa, F.M.; Turley, H.; A Knowles, M.; Cranston, D.; Catto, J.W.F.; et al. Hypoxia regulates FGFR3 expression via HIF-1α and miR-100 and contributes to cell survival in non-muscle invasive bladder cancer. Br. J. Cancer 2013, 109, 50–59, doi:10.1038/bjc.2013.240.
- Zhu, ; He, C.; Deng, S.; Li, X.; Cui, S.; Zeng, Z.; Liu, M.; Zhao, S.; Chen, J.; Jin, Y.; et al. MiR-548an, Transcriptionally Downregulated by HIF1α/HDAC1, Suppresses Tumorigenesis of Pancreatic Cancer by Targeting Vimentin Expression. Mol. Cancer Ther. 2016, 15, 2209–2219, doi:10.1158/1535-7163.mct-15-0877.
- He, ; Wang, Q.-Y.; Yin, Q.-Q.; Tang, J.; Lu, Y.; Zhou, C.-X.; Duan, C.-W.; Hong, D.-L.; Tanaka, T.; Chen, G.-Q.; et al. HIF-1α downregulates miR-17/20a directly targeting p21 and STAT3: A role in myeloid leukemic cell differentiation. Cell Death Differ. 2012, 20, 408–418, doi:10.1038/cdd.2012.130.
- Dico, L.; Costa, V.; Martelli, C.; Diceglie, C.; Rajata, F.; Rizzo, A.; Mancone, C.; Tripodi, M.; Ottobrini, L.; Alessandro, R.; et al. MiR675-5p Acts on HIF-1α to Sustain Hypoxic Responses: A New Therapeutic Strategy for Glioma. Theranostics 2016, 6, 1105–1118, doi:10.7150/thno.14700.
- Shi, ; Wang, Y.; Luan, W.; Wang, P.; Tao, T.; Zhang, J.; Qian, J.; Liu, N.; You, Y. Long Non-Coding RNA H19 Promotes Glioma Cell Invasion by Deriving miR-675. PLoS ONE 2014, 9, e86295, doi:10.1371/journal.pone.0086295.
- Costa, ; Dico, A.L.; Rizzo, A.; Rajata, F.; Tripodi, M.; Alessandro, R.; Conigliaro, A. MiR-675-5p supports hypoxia induced epithelial to mesenchymal transition in colon cancer cells. Oncotarget 2017, 8, 24292–24302, doi:10.18632/oncotarget.14464.
- Saieva, ; Barreca, M.M.; Zichittella, C.; Prado, M.G.; Tripodi, M.; Alessandro, R.; Conigliaro, A. Hypoxia-Induced miR-675-5p Supports β-Catenin Nuclear Localization by Regulating GSK3-β Activity in Colorectal Cancer Cell Lines. Int. J. Mol. Sci. 2020, 21, 3832, doi:10.3390/ijms21113832.
- Zheng, ; Wu, D.; Fan, S.; Zhang, Z.; Chen, G.; Lu, J. Upregulation of miR‐675‐5p induced by lncRNA H19 was associated with tumor progression and development by targeting tumor suppressor p53 in non–small cell lung cancer. J. Cell. Biochem. 2019, 120, 18724–18735, doi:10.1002/jcb.29182.
- Ge, ; Liu, X.; Lin, F.; Li, P.; Liu, K.; Geng, R.; Dai, C.; Lin, Y.; Tang, W.; Wu, Z.; et al. MicroRNA-421 regulated by HIF-1α promotes metastasis, inhibits apoptosis, and induces cisplatin resistance by targeting E-cadherin and caspase-3 in gastric cancer. Oncotarget 2016, 7, 24466–24482, doi:10.18632/oncotarget.8228.
- Liu, ; Wang, Y.; Dou, C.; Xu, M.; Sun, L.; Wang, L.; Yao, B.; Li, Q.; Yang, W.; Tu, K.; et al. Hypoxia-induced up-regulation of VASP promotes invasiveness and metastasis of hepatocellular carcinoma. Theranostics 2018, 8, 4649–4663, doi:10.7150/thno.26789.
- Guo, F.; Wang, A.Y.; Liu, J. HIFs-MiR-33a-Twsit1 axis can regulate invasiveness of hepatocellular cancer cells. Eur. Rev. Med. Pharmacol. Sci. 2016, 20, 3011–3016, doi:11171 [pii].
- Li, ; He, L.; Zuo, D.; He, W.; Wang, Y.; Zhang, Y.; Liu, W.; Yuan, Y. Mutual Regulation of MiR-199a-5p and HIF-1α Modulates the Warburg Effect in Hepatocellular Carcinoma. J. Cancer 2017, 8, 940–949, doi:10.7150/jca.17496.
- Huang, ; Ding, L.; Bennewith, K.L.; Tong, R.T.; Welford, S.M.; Ang, K.K.; Story, M.; Le, Q.-T.; Giaccia, A.J. Hypoxia-Inducible mir-210 Regulates Normoxic Gene Expression Involved in Tumor Initiation. Mol. Cell 2009, 35, 856–867, doi:10.1016/j.molcel.2009.09.006.
- Pasculli, ; Barbano, R.; Rendina, M.; Fontana, A.; Copetti, M.; Mazza, T.; Valori, V.M.; Morritti, M.; Maiello, E.; Graziano, P.; et al. Hsa-miR-210-3p expression in breast cancer and its putative association with worse outcome in patients treated with Docetaxel. Sci. Rep. 2019, 9, 1–9, doi:10.1038/s41598-019-51581-3.
- Osugi, ; Kimura, Y.; Owada, Y.; Inoue, T.; Watanabe, Y.; Yamaura, T.; Fukuhara, M.; Muto, S.; Okabe, N.; Matsumura, Y.; et al. Prognostic Impact of Hypoxia-Inducible miRNA-210 in Patients with Lung Adenocarcinoma. J. Oncol. 2015, 2015, 316745–8, doi:10.1155/2015/316745.
- Qu, ; Huang, W. Effects of microRNA‑210 on the diagnosis and treatment of prostate cancer. Mol. Med. Rep. 2018, 18, 1740–1744, doi:10.3892/mmr.2018.9105.
- Grosso, ; Doyen, J.; Parks, S.K.; Bertero, T.; Paye, A.; Cardinaud, B.; Gounon, P.; Lacas-Gervais, S.; Noël, A.; Pouysségur, J.; et al. MiR-210 promotes a hypoxic phenotype and increases radioresistance in human lung cancer cell lines. Cell Death Dis. 2013, 4, e544, doi:10.1038/cddis.2013.71.
- Devlin, ; Greco, S.; Martelli, F.; Ivan, M. miR-210: More than a silent player in hypoxia. IUBMB Life 2011, 63, 94–100, doi:10.1002/iub.427.
- Sahraei, ; Chaube, B.; Liu, Y.; Sun, J.; Kaplan, A.; Price, N.L.; Ding, W.; Oyaghire, S.; García-Milian, R.; Mehta, S.; et al. Suppressing miR-21 activity in tumor-associated macrophages promotes an antitumor immune response. J. Clin. Investig. 2019, 129, 5518–5536, doi:10.1172/jci127125.
- Li, ; Li, C.; Wang, S.; Wang, Z.; Jiang, J.; Wang, W.; Li, X.; Chen, J.; Liu, K.; Li, C.; et al. Exosomes Derived from Hypoxic Oral Squamous Cell Carcinoma Cells Deliver miR-21 to Normoxic Cells to Elicit a Prometastatic Phenotype. Cancer Res. 2016, 76, 1770–1780, doi:10.1158/0008-5472.can-15-1625.
- Ullmann, P.; Nurmik, M.; Schmitz, M.; Rodriguez, F.; Weiler, J.; Qureshi-Baig, K.; Felten, P.; Nazarov, P.V.; Nicot, N.; Zuegel, N.; et al. Tumor suppressor miR-215 counteracts hypoxia-induced colon cancer stem cell activity. Cancer Lett. 2019, 450, 32–41, doi:10.1016/j.canlet.2019.02.030.
References
- Camps, ; Saini, H.K.; Mole, D.R.; Choudhry, H.; Reczko, M.; Guerra-Assunção, J.A.; Tian, Y.-M.; Buffa, F.M.; Harris, A.L.; Hatzigeorgiou, A.G.; et al. Integrated analysis of microRNA and mRNA expression and association with HIF binding reveals the complexity of microRNA expression regulation under hypoxia. Mol. Cancer 2014, 13, 28, doi:10.1186/1476-4598-13-28.
- Nallamshetty, ; Chan, S.Y.; Loscalzo, J. Hypoxia: A master regulator of microRNA biogenesis and activity. Free Radic. Biol. Med. 2013, 64, 20–30, doi:10.1016/j.freeradbiomed.2013.05.022.
- Slemc, ; Kunej, T. Transcription factor HIF1A: Downstream targets, associated pathways, polymorphic hypoxia response element (HRE) sites, and initiative for standardization of reporting in scientific literature. Tumor Biol. 2016, 37, 14851–14861, doi:10.1007/s13277-016-5331-4.
- Choudhry, ; Schödel, J.; Oikonomopoulos, S.; Camps, C.; Grampp, S.; Harris, A.L.; Ratcliffe, P.J.; Ragoussis, J.; Mole, D.R. Extensive regulation of the non‐coding transcriptome by hypoxia: Role of HIF in releasing paused RNA pol2. EMBO Rep. 2014, 15, 70–76, doi:10.1002/embr.201337642.
- Lai, -H.; Li, J.-N.; Wang, M.-Y.; Huang, H.-Y.; Croce, C.M.; Sun, H.-L.; Lyu, Y.-J.; Kang, J.-W.; Chiu, C.-F.; Hung, M.-C.; et al. HIF-1α promotes autophagic proteolysis of Dicer and enhances tumor metastasis. J. Clin. Investig. 2017, 128, 625–643, doi:10.1172/jci89212.
- Wang, ; Liu, X.; Zhang, H.; Sun, L.; Zhou, Y.; Jin, H.; Zhang, H.; Zhang, H.; Liu, J.; Guo, H.; et al. Hypoxia-inducible lncRNA-AK058003 promotes gastric cancer metastasis by targeting γ-synuclein. Neoplasia 2014, 16, 1094–1106, doi:10.1016/j.neo.2014.10.008.
- He, ; Wang, P. Unregulated long non-coding RNA-AK058003 promotes the proliferation, invasion and metastasis of breast cancer by regulating the expression levels of the γ-synuclein gene. Exp. Ther. Med. 2015, 9, 1727–1732, doi:10.3892/etm.2015.2323.
- Cheng, ; Qiu, J.; Wang, S.; Yang, Y.; Guo, M.; Wang, D.; Luo, Q.; Xu, L. Comprehensive circular RNA profiling identifies CircFAM120A as a new biomarker of hypoxic lung adenocarcinoma. Ann. Transl. Med. 2019, 7, 442, doi:10.21037/atm.2019.08.79.
- Di Liddo, ; Machado, C.D.O.F.; Fischer, S.; Ebersberger, S.; Heumüller, A.W.; E Weigand, J.; Müller-McNicoll, M.; Zarnack, K. A combined computational pipeline to detect circular RNAs in human cancer cells under hypoxic stress. J. Mol. Cell Biol. 2019, 11, 829–844, doi:10.1093/jmcb/mjz094.
- Wei, ; Zhang, Y.; Meng, Q.; Cui, L.; Xu, C. Hypoxia-induced circular RNA has_circRNA_403658 promotes bladder cancer cell growth through activation of LDHA. Am. J. Transl Res. 2019, 11, 6838–6849.
- Wang, ; Deng, M.; Liu, Z.; Wu, S. Hypoxia-induced miR-210 promoter demethylation enhances proliferation, autophagy and angiogenesis of schwannoma cells. Oncol. Rep. 2017, 37, 3010–3018, doi:10.3892/or.2017.5511.
- Chio, -C.; Lin, J.-W.; Cheng, H.-A.; Chiu, W.-T.; Wang, Y.-H.; Wang, J.-J.; Hsing, C.-H.; Chen, R.-M. MicroRNA-210 targets antiapoptotic Bcl-2 expression and mediates hypoxia-induced apoptosis of neuroblastoma cells. Arch. Toxicol. 2012, 87, 459–468, doi:10.1007/s00204-012-0965-5.
- Zhang, ; Sun, H.; Dai, H.; Walsh, R.M.; Imakura, M.; Schelter, J.; Burchard, J.; Dai, X.; Chang, A.N.; Diaz, R.L.; et al. MicroRNA miR-210 modulates cellular response to hypoxia through the MYC antagonist MNT. Cell Cycle 2009, 8, 2756–2768, doi:10.4161/cc.8.17.9387.
- Yang, ; Wei, J.; Guo, T.; Shen, Y.; Liu, F. Knockdown of miR-210 decreases hypoxic glioma stem cells stemness and radioresistance. Exp. Cell Res. 2014, 326, 22–35, doi:10.1016/j.yexcr.2014.05.022.
- Li, ; Huang, K.; You, Y.; Fu, X.; Hu, L.; Song, L.; Meng, Y. Hypoxia-induced miR-210 in epithelial ovarian cancer enhances cancer cell viability via promoting proliferation and inhibiting apoptosis. Int. J. Oncol. 2014, 44, 2111–2120, doi:10.3892/ijo.2014.2368.
- Yang, ; Sun, T.; Cao, J.; Liu, F.; Tian, Y.; Zhu, W. Downregulation of miR-210 expression inhibits proliferation, induces apoptosis and enhances radiosensitivity in hypoxic human hepatoma cells in vitro. Exp. Cell Res. 2012, 318, 944–954, doi:10.1016/j.yexcr.2012.02.010.
- Xue, -X.; Shang, C.; Hong, Y.; Guo, Y.; Liu, Y.-H. MiR-210 Up-Regulation Inhibits Proliferation and Induces Apoptosis in Glioma Cells by Targeting SIN3A. Med. Sci. Monit. 2014, 20, 2571–2577, doi:10.12659/msm.892994.
- Du, ; Wei, N.; Ma, R.; Jiang, S.; Song, D. A miR-210-3p regulon that controls the Warburg effect by modulating HIF-1α and p53 activity in triple-negative breast cancer. Cell Death Dis. 2020, 11, 1–12, doi:10.1038/s41419-020-02952-6.
- Blick, ; Ramachandran, A.V.; McCormick, R.; Wigfield, S.; Cranston, D.; Catto, J.W.F.; Harris, A.L. Identification of a hypoxia-regulated miRNA signature in bladder cancer and a role for miR-145 in hypoxia-dependent apoptosis. Br. J. Cancer 2015, 113, 634–644, doi:10.1038/bjc.2015.203.
- Nagpal, ; Ahmad, H.M.; Chameettachal, S.; Sundar, D.; Ghosh, S.; Kulshreshtha, R. HIF-inducible miR-191 promotes migration in breast cancer through complex regulation of TGFβ-signaling in hypoxic microenvironment. Sci. Rep. 2015, 5, 9650, doi:10.1038/srep09650.
- Zhao, ; Qiao, C.-R.; Ding, Z.; Sheng, Y.-L.; Li, X.-N.; Yang, Y.; Zhu, D.-Y.; Zhang, C.-Y.; Liu, D.-L.; Wu, K.; et al. A novel pathway in NSCLC cells: miR-191, targeting NFIA, is induced by chronic hypoxia, and promotes cell proliferation and migration. Mol. Med. Rep. 2017, 15, 1319–1325, doi:10.3892/mmr.2017.6100.
- Zhao, ; Li, Y.; Tan, B.-B.; Fan, L.-Q.; Yang, P.-G.; Tian, Y. HIF-1α Induces Multidrug Resistance in Gastric Cancer Cells by Inducing MiR-27a. PLoS ONE 2015, 10, e0132746, doi:10.1371/journal.pone.0132746.
- Seok, -K.; Lee, S.H.; Kim, M.J.; Lee, Y.-M. MicroRNA-382 induced by HIF-1α is an angiogenic miR targeting the tumor suppressor phosphatase and tensin homolog. Nucleic Acids Res. 2014, 42, 8062–8072, doi:10.1093/nar/gku515.
- Zhang, ; Shi, Z.; Li, M.; Mi, J. Hypoxia-induced miR-424 decreases tumor sensitivity to chemotherapy by inhibiting apoptosis. Cell Death Dis. 2014, 5, e1301, doi:10.1038/cddis.2014.240.
- Chen, -Y.; Lin, Y.-M.; Chung, H.-C.; Lang, Y.-D.; Lin, C.-J.; Huang, J.; Wang, W.-C.; Lin, F.-M.; Chen, Z.; Huang, H.-D.; et al. miR-103/107 Promote Metastasis of Colorectal Cancer by Targeting the Metastasis Suppressors DAPK and KLF4. Cancer Res. 2012, 72, 3631–3641, doi:10.1158/0008-5472.can-12-0667.
- Mace, A.; Collins, A.L.; Wojcik, S.E.; Croce, C.M.; Lesinski, G.B.; Bloomston, M. Hypoxia induces the overexpression of microRNA-21 in pancreatic cancer cells. J. Surg. Res. 2013, 184, 855–860, doi:10.1016/j.jss.2013.04.061.
- Song, ; Liu, S.; Zhang, L.; Yao, H.; Gao, F.; Xu, D.; Lili, S. MiR-21 modulates radiosensitivity of cervical cancer through inhibiting autophagy via the PTEN/Akt/HIF-1α feedback loop and the Akt-mTOR signaling pathway. Tumor Biol. 2016, 37, 12161–12168, doi:10.1007/s13277-016-5073-3.
- He, ; Wang, L.; Zhang, J.; Xu, H. Hypoxia-inducible microRNA-224 promotes the cell growth, migration and invasion by directly targeting RASSF8 in gastric cancer. Mol. Cancer 2017, 16, 1–14, doi:10.1186/s12943-017-0603-1.
- Blick, ; Ramachandran, A.; Wigfield, S.; McCormick, R.; Jubb, A.M.; Buffa, F.M.; Turley, H.; A Knowles, M.; Cranston, D.; Catto, J.W.F.; et al. Hypoxia regulates FGFR3 expression via HIF-1α and miR-100 and contributes to cell survival in non-muscle invasive bladder cancer. Br. J. Cancer 2013, 109, 50–59, doi:10.1038/bjc.2013.240.
- Zhu, ; He, C.; Deng, S.; Li, X.; Cui, S.; Zeng, Z.; Liu, M.; Zhao, S.; Chen, J.; Jin, Y.; et al. MiR-548an, Transcriptionally Downregulated by HIF1α/HDAC1, Suppresses Tumorigenesis of Pancreatic Cancer by Targeting Vimentin Expression. Mol. Cancer Ther. 2016, 15, 2209–2219, doi:10.1158/1535-7163.mct-15-0877.
- He, ; Wang, Q.-Y.; Yin, Q.-Q.; Tang, J.; Lu, Y.; Zhou, C.-X.; Duan, C.-W.; Hong, D.-L.; Tanaka, T.; Chen, G.-Q.; et al. HIF-1α downregulates miR-17/20a directly targeting p21 and STAT3: A role in myeloid leukemic cell differentiation. Cell Death Differ. 2012, 20, 408–418, doi:10.1038/cdd.2012.130.
- Dico, L.; Costa, V.; Martelli, C.; Diceglie, C.; Rajata, F.; Rizzo, A.; Mancone, C.; Tripodi, M.; Ottobrini, L.; Alessandro, R.; et al. MiR675-5p Acts on HIF-1α to Sustain Hypoxic Responses: A New Therapeutic Strategy for Glioma. Theranostics 2016, 6, 1105–1118, doi:10.7150/thno.14700.
- Shi, ; Wang, Y.; Luan, W.; Wang, P.; Tao, T.; Zhang, J.; Qian, J.; Liu, N.; You, Y. Long Non-Coding RNA H19 Promotes Glioma Cell Invasion by Deriving miR-675. PLoS ONE 2014, 9, e86295, doi:10.1371/journal.pone.0086295.
- Costa, ; Dico, A.L.; Rizzo, A.; Rajata, F.; Tripodi, M.; Alessandro, R.; Conigliaro, A. MiR-675-5p supports hypoxia induced epithelial to mesenchymal transition in colon cancer cells. Oncotarget 2017, 8, 24292–24302, doi:10.18632/oncotarget.14464.
- Saieva, ; Barreca, M.M.; Zichittella, C.; Prado, M.G.; Tripodi, M.; Alessandro, R.; Conigliaro, A. Hypoxia-Induced miR-675-5p Supports β-Catenin Nuclear Localization by Regulating GSK3-β Activity in Colorectal Cancer Cell Lines. Int. J. Mol. Sci. 2020, 21, 3832, doi:10.3390/ijms21113832.
- Zheng, ; Wu, D.; Fan, S.; Zhang, Z.; Chen, G.; Lu, J. Upregulation of miR‐675‐5p induced by lncRNA H19 was associated with tumor progression and development by targeting tumor suppressor p53 in non–small cell lung cancer. J. Cell. Biochem. 2019, 120, 18724–18735, doi:10.1002/jcb.29182.
- Ge, ; Liu, X.; Lin, F.; Li, P.; Liu, K.; Geng, R.; Dai, C.; Lin, Y.; Tang, W.; Wu, Z.; et al. MicroRNA-421 regulated by HIF-1α promotes metastasis, inhibits apoptosis, and induces cisplatin resistance by targeting E-cadherin and caspase-3 in gastric cancer. Oncotarget 2016, 7, 24466–24482, doi:10.18632/oncotarget.8228.
- Liu, ; Wang, Y.; Dou, C.; Xu, M.; Sun, L.; Wang, L.; Yao, B.; Li, Q.; Yang, W.; Tu, K.; et al. Hypoxia-induced up-regulation of VASP promotes invasiveness and metastasis of hepatocellular carcinoma. Theranostics 2018, 8, 4649–4663, doi:10.7150/thno.26789.
- Guo, F.; Wang, A.Y.; Liu, J. HIFs-MiR-33a-Twsit1 axis can regulate invasiveness of hepatocellular cancer cells. Eur. Rev. Med. Pharmacol. Sci. 2016, 20, 3011–3016, doi:11171 [pii].
- Li, ; He, L.; Zuo, D.; He, W.; Wang, Y.; Zhang, Y.; Liu, W.; Yuan, Y. Mutual Regulation of MiR-199a-5p and HIF-1α Modulates the Warburg Effect in Hepatocellular Carcinoma. J. Cancer 2017, 8, 940–949, doi:10.7150/jca.17496.
- Huang, ; Ding, L.; Bennewith, K.L.; Tong, R.T.; Welford, S.M.; Ang, K.K.; Story, M.; Le, Q.-T.; Giaccia, A.J. Hypoxia-Inducible mir-210 Regulates Normoxic Gene Expression Involved in Tumor Initiation. Mol. Cell 2009, 35, 856–867, doi:10.1016/j.molcel.2009.09.006.
- Pasculli, ; Barbano, R.; Rendina, M.; Fontana, A.; Copetti, M.; Mazza, T.; Valori, V.M.; Morritti, M.; Maiello, E.; Graziano, P.; et al. Hsa-miR-210-3p expression in breast cancer and its putative association with worse outcome in patients treated with Docetaxel. Sci. Rep. 2019, 9, 1–9, doi:10.1038/s41598-019-51581-3.
- Qu, ; Huang, W. Effects of microRNA‑210 on the diagnosis and treatment of prostate cancer. Mol. Med. Rep. 2018, 18, 1740–1744, doi:10.3892/mmr.2018.9105.
- Grosso, ; Doyen, J.; Parks, S.K.; Bertero, T.; Paye, A.; Cardinaud, B.; Gounon, P.; Lacas-Gervais, S.; Noël, A.; Pouysségur, J.; et al. MiR-210 promotes a hypoxic phenotype and increases radioresistance in human lung cancer cell lines. Cell Death Dis. 2013, 4, e544, doi:10.1038/cddis.2013.71.
- Devlin, ; Greco, S.; Martelli, F.; Ivan, M. miR-210: More than a silent player in hypoxia. IUBMB Life 2011, 63, 94–100, doi:10.1002/iub.427.
- Sahraei, ; Chaube, B.; Liu, Y.; Sun, J.; Kaplan, A.; Price, N.L.; Ding, W.; Oyaghire, S.; García-Milian, R.; Mehta, S.; et al. Suppressing miR-21 activity in tumor-associated macrophages promotes an antitumor immune response. J. Clin. Investig. 2019, 129, 5518–5536, doi:10.1172/jci127125.
- Li, ; Li, C.; Wang, S.; Wang, Z.; Jiang, J.; Wang, W.; Li, X.; Chen, J.; Liu, K.; Li, C.; et al. Exosomes Derived from Hypoxic Oral Squamous Cell Carcinoma Cells Deliver miR-21 to Normoxic Cells to Elicit a Prometastatic Phenotype. Cancer Res. 2016, 76, 1770–1780, doi:10.1158/0008-5472.can-15-1625.
- Ullmann, P.; Nurmik, M.; Schmitz, M.; Rodriguez, F.; Weiler, J.; Qureshi-Baig, K.; Felten, P.; Nazarov, P.V.; Nicot, N.; Zuegel, N.; et al. Tumor suppressor miR-215 counteracts hypoxia-induced colon cancer stem cell activity. Cancer Lett. 2019, 450, 32–41, doi:10.1016/j.canlet.2019.02.030.