Plant NBR1 is a structural homolog and functional hybrid of mammalian autophagy receptors NBR1 and p62.
- autophagy
- selective autophagy receptors
- plants stress responses
- NBR1
1. Introduction
Autophagy is a highly conserved pathway in eukaryotes that recycles multiple cytoplasmic components under both normal and stress conditions such as starvation [1]. Induction of autophagy is initiated by the formation of an isolation membrane called phagophore that can extend to capture and sequester cytoplasmic components within a double-membrane vesicle termed autophagosome [2,3][2][3]. Mature autophagosomes can then fuse with the lysosomes or vacuoles for degradation of their cargo by resident hydrolases. The core machinery of autophagosome formation requires more than 40 largely conserved autophagy-related proteins (ATG). In vertebrates, these core autophagy components function in several physiologically continuous, but mechanistically distinct, steps and are organized into several functional complexes including: (i) the ULK (Unc-51 Like Autophagy Activating Kinase) complex with ULK1 and -2, ATG13, ATG101 and FIP200 (FAK Family Kinase-Interacting Protein of 200 kDa), (ii) the class III phosphoinositide 3-kinase (PI3K) complex I, with VPS34 (Vacuolar Protein Sorting 34), VPS15, Beclin1 and ATG14 for the nucleation and assembly of the initial phagophore membrane, (iii) the phosphatidylinositol-3-phosphate (PI3P)-binding ATG2A or -B and WIPI1–4 (WD Repeat Domain Phosphoinositide-Interacting Protein 1–4) complex, and iv) the two interrelated ubiquitin-like conjugation systems, ATG12–ATG5–ATG16 and ATG8–PE (phosphatidylethanolamine), which are required for the membrane elongation and expansion of the forming autophagosomes [4]. In addition, the ATG4 cysteine proteases process the precursors of ATG8 proteins for their lipidation and delipidation [4].
During cellular response to nutrient deprivation, autophagy usually involves non-selective uptake of cytoplasm into phagophores for bulk degradation of intracellular contents [5]. However, the broad roles of autophagy are primarily mediated by selective clearance of certain components [5]. In human cells, extensive studies have reported the selective autophagic degradation of aggregation-prone misfolded proteins and protein aggregates implicated in the pathology of various neurodegenerative diseases [5]. Furthermore, autophagy selectively degrades diverse organelles such as mitochondria, peroxisomes, lysosomes, endoplasmic reticulum (ER) and the nucleus, under various conditions [5]. Ubiquitin-like ATG8 plays a critical role in selective autophagy [6]. After attachment of the lipid PE to its carboxyl terminus through a conjugation pathway, ATG8 is both anchored in the membrane of autophagosomes and acts as a docking platform for the selective recruitment of cargos through a three-way interaction of selective autophagy receptors with both ATG8 and cargos [6]. Most selective autophagy receptors interact with membrane-anchored ATG8 through ATG8-interacting motifs (AIMs), which have the W/Y/F-X-X-L/I/V consensus core sequence [6]. AIMs of selective autophagy receptors bind a hydrophobic patch on ATG8 known as the AIM docking site [7]. A new class of selective autophagy receptors have been recently identified that interact with ATG8 through ubiquitin-interacting motif (UIM)-like sequences for high-affinity binding to an ATG8 interaction site different from the AIM docking site [8,9][8][9].
Autophagy has been extensively analyzed over the past two decades or so in Arabidopsis and, to a lesser extent, in other plants. Using genetic and molecular approaches, these extensive studies have established an important role of autophagy in almost all aspects of plant life, particularly in plant stress responses [10,11][10][11]. Autophagosome biogenesis and ATG gene expression are both induced under diverse abiotic stress conditions including nutrient starvation, heat, salt, drought and oxidative stresses [12,13,14,15,16,17][12][13][14][15][16][17]. Autophagy mutants and transgenic silencing lines display increased sensitivity to nutrient starvation and abiotic stresses when compared to wild-type plants [12,13,14,15,16,17][12][13][14][15][16][17]. In addition, plant mutants or transgenic silencing lines for autophagy are altered in response to virulent and avirulent biotrophic pathogens including pathogen-induced hypersensitive cell death [18,19,20,21,22,23][18][19][20][21][22][23]. Autophagy-deficient mutants are hypersusceptible to necrotrophic pathogens [19,24][19][24]. Furthermore, autophagy affects plant interaction with viral pathogens through regulation of antiviral RNA silencing, targeting degradation of viral proteins and other processes [20,25,26,27,28][20][25][26][27][28]. Autophagy also plays important roles in plant growth and development including root growth, leaf senescence, pollen and endosperm development [22,29,30,31,32][22][29][30][31][32].
Over the past ten years or so, a substantial number of selective autophagy receptors have been identified, characterized and functionally analyzed in plants [33][34] [33,34] (Table 1; Figure 1). While a few of these autophagy receptors in plants are evolutionarily conserved with homologs in other types of organisms, most of them are plant-specific or even plant species-specific. The cargos recognized by these plant selective autophagy receptors include not only misfolded, nonactive and otherwise unwanted cellular components, but also regulatory and signaling factors [33,34][33][34]. Characterization of these plant autophagy receptors and their cargos have provided important new insights into the critical roles of autophagy in plant responses to a broad spectrum of biotic and abiotic stresses. Several recent reviews have covered selective autophagy in plants that also include discussion on well-studied selective autophagy receptors from plants [34,35,36][34][35][36].
.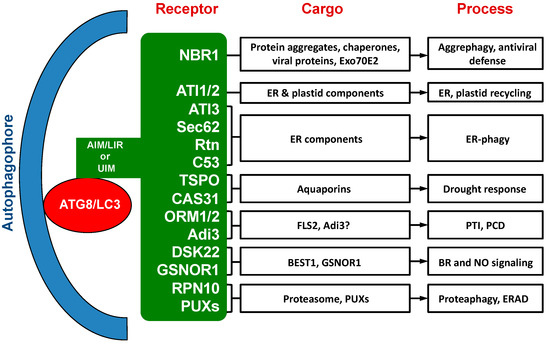 Figure 1. Cargo proteins and involved processes of selective autophagy receptors in plants.
Figure 1. Cargo proteins and involved processes of selective autophagy receptors in plants.
Table 1. Interacting ATG8 (autophagy-related protein 8) isoforms and recognition motifs of selective autophagy from plants.
| Receptor | Interacting ATG8 Isoform | Recognition Motif |
|---|---|---|
| NBR1 | ATG8s | AIM |
| ATI1/2 | ATG8f, ATG8h | AIM |
| ATI3s | ATG8a, ATG8f | AIM |
| Sec62 | ATG8e | AIM |
| ZmRtn1/2 | ZmATG8a | AIM |
| C53 | ATG8a–g, ATG8i | AIM |
| TSPO | ATG8e | AIM |
| MtCAS31 | MtATG8a | AIM |
| ORM1/2 | ATG8a, ATG8d, ATG8e, ATG8i | AIM |
| SlAdi3 | SlATG8h | AIM |
| DSK2 | ATG8e | AIM |
| GSNOR1 | ATG8s | AIM |
| RPN10 | ATGa, ATG8e, ATG8f, ATG8i | UIM |
| PUXs | ATG8a, ATG8e | UIM |
2. NBR1 (Neighbor of BRCA1)
Among the identified autophagy receptors in plants, NBR1 has been most extensively characterized. Plant NBR1 is a structural homolog and functional hybrid of mammalian autophagy receptors NBR1 and p62 [51,52][51][52]. Both mammalian p62 and NBR1 proteins contain an N-terminal PB1 (Phox and Bem1p) domain, a ZZ-type zinc finger domain, an LC3-interacting region or LIR motif (also known as AIM motif in yeast and plants) and a C-terminal UBA (ubiquitin-associated) domain [53]. In addition, there is a highly conserved globular domain characterized by the presence of four highly conserved tryptophan residues in NBR1 but not in p62 [53]. Only metazoans contain both p62 and NBR1 homologs, while other eukaryotic organisms only have NBR homologs [53]. Plant NBR1 homologs lack the coiled coil domain of mammalian NBR1 but have two C-terminal UBA domains [37]. Model plant Arabidopsis contains a single gene encoding an NBR1 homolog, which, however, can homo-oligomerize through the N-terminal PB1 domain like p62 [37]. Only the C-terminal UBA domain of the two UBA domains of Arabidopsis NBR1 binds ubiquitin [37].
The biological functions of plant NBR1 have been analyzed through characterization of nbr1 mutants or transgenic silencing lines. Arabidopsis nbr1 knockout mutants are normal in growth and development under normal growth conditions. The nbr1 mutants are also normal in general autophagy and in the selective clearance of peroxisomes, mitochondria, or the endoplasmic reticulum (ER) [16,54,55,56][16][54][55][56]. Plant NBR1 is not essential either for age- and darkness-induced senescence but may modulate growth or senescence under certain conditions such as short-day growth condition or under mineral deficiency [16,52,57][16][52][57]. The Arabidopsis nbr1 mutants also respond normally to a necrotrophic pathogen [16]. However, loss of Arabidopsis NBR1 gene function compromise plant tolerance to heat, oxidative, salt, and drought stresses [16,55][16][55]. The role of NBR1 in plant abiotic stress tolerance is mediated by selective autophagy based on its dependent on the interaction with ATG8 and is associated with the clearance of aggregation-prone misfolded proteins and protein aggregates [16] [16] (Figure 1). In Arabidopsis, NBR1 also plays a role in resistance to the bacterial pathogen Pseudomonas syringae by suppressing the establishment of an aqueous extracellular space (“water-soaking”) [58]. More recent studies have further revealed important roles of plant NBR1 in the modulation of plant heat stress memory, plant–viral interaction and other stress-associated processes. These new roles of NBR1-mediated selective autophagy will be discussed in more detail here.Very often, plants can be subjected repeatedly to a stress condition such as high temperature and need to balance between growth recovery and keeping stress memory for better survival when faced with a subsequent harsher stress [59]. Autophagy is induced in plants by moderate heat stress and targets a large number of proteins including specific heat shock proteins (HSPs) for degradation during the recovery phase after the end of heat stress, leading to reduced heat stress memory [59]. These target proteins include HSP90.1 and its interacting partner ROF1/AFKBP62 (rotamase FKBP 1), a plant homolog of mammalian FKBP4/FKBP52 [16,60][16][60]. The HSP90.1-ROF1 complex remains in the cytoplasm under normal conditions but binds heat shock transcription factor HSFA2 and translocates to the nucleus to activate heat-responsive gene expression following exposure to heat stress [61]. Degradation of HSP90.1 and ROF1 by NBR1-mediated selective autophagy attenuates HSFA2-dependent induction of HSP genes and represses the response to heat stress [60]. Indeed, the nbr1 loss-of-function mutants is stronger in heat stress memory [60]. These results indicate that plant NBR1 plays complex roles in plant heat stress responses. It promotes basal heat tolerance mostly through autophagic degradation of misfolded/denatured proteins or protein aggregates to mitigate heat-induced proteotoxicity (Figure 2). After the end of heat stress, NBR1-mediated selective autophagy targets degradation of specific HSPs to reduce heat stress memory, probably to promote growth recovery but also downregulate acquired heat tolerance to a potential subsequent heat stress (Figure 2).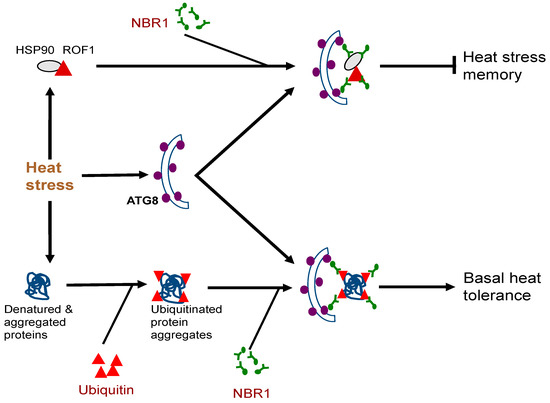 Figure 2. Complex roles of plant NBR1 (Neighbor of BRCA1) in plant heat stress responses. It mediates autophagic degradation of misfolded/denatured proteins or protein aggregates to mitigate heat-induced proteotoxicity to promote basal heat tolerance. NBR1-mediated selective autophagy also targets degradation of specific heat shock proteins (HSPs) such as HSP90 to reduce heat stress memory.
Figure 2. Complex roles of plant NBR1 (Neighbor of BRCA1) in plant heat stress responses. It mediates autophagic degradation of misfolded/denatured proteins or protein aggregates to mitigate heat-induced proteotoxicity to promote basal heat tolerance. NBR1-mediated selective autophagy also targets degradation of specific heat shock proteins (HSPs) such as HSP90 to reduce heat stress memory.
Autophagy plays an important role in plant–virus interactions. Previous studies have demonstrated that autophagy regulates virus-induced hypersensitive cell death and targets degradation of plant and viral proteins associated with dsRNA-induced RNA silencing [20,25][20][25]. More recent studies have revealed that NBR1-mediated selective autophagy targets degradation of specific viral proteins to suppress viral infection. In the study with Cauliflower mosaic virus (CaMV), it has been shown that NBR1-mediated selective autophagy targets nonassembled and virus particle-forming capsid proteins for degradation to restrict the establishment of CaMV infection [26]. To counter the antiviral defense mechanism, the CaMV-induced virus factory inclusions sequester the viral proteins and coordinate particle assembly and storage to protect capsid proteins against autophagic destruction [26][59]. NBR1 also targets the viral RNA silencing suppressor helper-component proteinase (HCpro), presumably in association with virus-induced RNA granules, to suppress accumulation of Turnip mosaic virus (TuMV), a positive-stranded RNA potyvirus [27]. Again, as counter defense mechanisms, several viral proteins have evolved the activity to antagonize NBR1-dependent autophagy. These results demonstrate the critical role of NBR1-mediated selective autophagy in plant antiviral defense and the potential viral strategies to evade and adapt autophagic processes for successful infection.Recent studies have also demonstrated other cargos recognized and potentially targeted by NBR1 and further illustrate a broad role of the autophagy receptor in plant metabolism and stress responses. For example, Arabidopsis NBR1 is a selective receptor for Exo70E2 during autophagy in Arabidopsis [57]. Exo70E2 is a subunit of the exocyst complex, which directs the secretory vesicles of exocytosis from the Golgi complex to specific locations on the plasma membrane and to mediate their tethering and localization to the membrane immediately before fusion [62]. In Arabidopsis, there is a double-membrane organelle termed the exocyst-positive organelle (EXPO), which may be involved in mediating unconventional protein secretion in plants [63,64][63][64]. Exo70E2 is a marker for EXPO [63]. Upon induction of autophagy, Exo70E2-GFP positive EXPOs and autophagosome were colocalized and delivered to vacuoles for degradation in transgenic Arabidopsis plants [63]. Arabidopsis NBR1 specifically interacted and recruited Exo70E2 or its EXPO to ATG8-positive autophagosomes in a manner independent of its UBA domains [57]. Knockout of the NBR1 gene significantly reduced the vacuolar delivery of Exo70E2 or EXPO upon autophagic induction [57] [57], supporting that the Arabidopsis NBR1-mediated selective autophagy pathway is involved in the vacuolar delivery of Exo70E2 or EXPO in plant autophagy.Arabidopsis NBR1 also interacts with members of the plant-specific LSU (response to Low SUlfur) protein family, which are induced by sulfur (S) deficiency, suggesting a possible role of NBR1 in plant S nutrient responses [65]. Indeed, S deficiency induces autophagy and the transcription of NBR1. NBR1 overexpression alters plant gene expression in response to the low S conditions [66]. Furthermore, Arabidopsis seedlings overexpressing NBR1 have significantly shorter roots than wild type when grown under S deficient conditions in the presence of TOR kinase inhibitors [66]. Arabidopsis NBR1 also interacts with three regulatory proteins of the abscisic acid (ABA) pathway (ABI3, ABI4 and ABI5) in planta [67]. NBR1 interaction with ABI5, but not ABI3 or ABI4, requires its UBA domain [67]. It is likely that ABI5, but not ABI3 or ABI4, requires ubiquitination prior to interaction with NBR1. It would be of interest to determine whether NBR1 binding of ABI3, ABI4 and ABI5 causes their autophagic degradation and affects their protein levels and ABA signaling in plants.
References
- Klionsky, D.J. Autophagy participates in, well, just about everything. Cell Death Differ. 2020, 27, 831–832, doi:10.1038/s41418-020-0511-6.
- Hansen, T.E.; Johansen, T. Following autophagy step by step. BMC Biol. 2011, 9, 39, doi:10.1186/1741-7007-9-39.
- Mizushima, N. Autophagy: Process and function. Genes Dev. 2007, 21, 2861–2873, doi:10.1101/gad.1599207.
- Yin, Z.Y.; Pascual, C.; Klionsky, D.J. Autophagy: Machinery and regulation. Microb. Cell 2016, 3, 457–465, doi:10.15698/mic2016.12.546.
- Gatica, D.; Lahiri, V.; Klionsky, D.J. Cargo recognition and degradation by selective autophagy. Nat. Cell Biol. 2018, 20, 233–242, doi:10.1038/s41556-018-0037-z.
- Abdrakhmanov, A.; Gogvadze, V.; Zhivotovsky, B. To Eat or to Die: Deciphering Selective Forms of Autophagy. Trends Biochem. Sci. 2020, 45, 347–364, doi:10.1016/j.tibs.2019.11.006.
- Johansen, T.; Lamark, T. Selective Autophagy: ATG8 Family Proteins, LIR Motifs and Cargo Receptors. J. Mol. Biol. 2020, 432, 80–103, doi:10.1016/j.jmb.2019.07.016.
- Marshall, R.S.; Hua, Z.H.; Mali, S.; McLoughlin, F.; Vierstra, R.D. ATG8-Binding UIM Proteins Define a New Class of Autophagy Adaptors and Receptors. Cell 2019, 177, 766–781, doi:10.1016/j.cell.2019.02.009.
- Marshall, R.S.; Li, F.; Gemperline, D.C.; Book, A.J.; Vierstra, R.D. Autophagic Degradation of the 26S Proteasome Is Mediated by the Dual ATG8/Ubiquitin Receptor RPN10 in Arabidopsis. Mol. Cell 2015, 58, 1053–1066, doi:10.1016/j.molcel.2015.04.023.
- Signorelli, S.; Tarkowski, L.P.; Van den Ende, W.; Bassham, D.C. Linking Autophagy to Abiotic and Biotic Stress Responses. Trends Plant Sci. 2019, 24, 413–430, doi:10.1016/j.tplants.2019.02.001.
- Tang, J.; Bassham, D.C. Autophagy in crop plants: what's new beyond Arabidopsis? Open Biol. 2018, 8, doi:10.1098/rsob.180162.
- Hanaoka, H.; Noda, T.; Shirano, Y.; Kato, T.; Hayashi, H.; Shibata, D.; Tabata, S.; Ohsumi, Y. Leaf senescence and starvation-induced chlorosis are accelerated by the disruption of an Arabidopsis autophagy gene. Plant Physiol. 2002, 129, 1181–1193, doi:10.1104/pp.011024.
- Liu, Y.; Xiong, Y.; Bassham, D.C. Autophagy is required for tolerance of drought and salt stress in plants. Autophagy 2009, 5, 954–963, doi:10.4161/auto.5.7.9290.
- Pu, Y.; Bassham, D.C. Links between ER stress and autophagy in plants. Plant Signal. Behav. 2013, 8, e24297, doi:10.4161/psb.24297.
- Wada, S.; Ishida, H.; Izumi, M.; Yoshimoto, K.; Ohsumi, Y.; Mae, T.; Makino, A. Autophagy plays a role in chloroplast degradation during senescence in individually darkened leaves. Plant Physiol. 2009, 149, 885–893, doi:10.1104/pp.108.130013.
- Zhou, J.; Wang, J.; Cheng, Y.; Chi, Y.J.; Fan, B.; Yu, J.Q.; Chen, Z. NBR1-mediated selective autophagy targets insoluble ubiquitinated protein aggregates in plant stress responses. PLoS Genet. 2013, 9, e1003196, doi:10.1371/journal.pgen.1003196.
- Zhou, J.; Wang, J.; Yu, J.Q.; Chen, Z. Role and regulation of autophagy in heat stress responses of tomato plants. Front. Plant Sci. 2014, 5, 174, doi:10.3389/fpls.2014.00174.
- Hayward, A.P.; Tsao, J.; Dinesh-Kumar, S.P. Autophagy and plant innate immunity: Defense through degradation. Semin. Cell Dev. Biol. 2009, 20, 1041–1047, doi:10.1016/j.semcdb.2009.04.012.
- Lenz, H.D.; Haller, E.; Melzer, E.; Gust, A.A.; Nurnberger, T. Autophagy controls plant basal immunity in a pathogenic lifestyle-dependent manner. Autophagy 2011, 7, 773–774, doi:10.4161/auto.7.7.15535.
- Liu, Y.; Schiff, M.; Czymmek, K.; Talloczy, Z.; Levine, B.; Dinesh-Kumar, S.P. Autophagy regulates programmed cell death during the plant innate immune response. Cell 2005, 121, 567–577, doi:10.1016/j.cell.2005.03.007.
- Wang, Y.; Wu, Y.; Tang, D. The autophagy gene, ATG18a, plays a negative role in powdery mildew resistance and mildew-induced cell death in Arabidopsis. Plant Signal. Behav. 2011, 6, 1408–1410, doi:10.4161/psb.6.9.16967.
- Yoshimoto, K.; Jikumaru, Y.; Kamiya, Y.; Kusano, M.; Consonni, C.; Panstruga, R.; Ohsumi, Y.; Shirasu, K. Autophagy negatively regulates cell death by controlling NPR1-dependent salicylic acid signaling during senescence and the innate immune response in Arabidopsis. Plant Cell 2009, 21, 2914–2927, doi:10.1105/tpc.109.068635.
- Zhou, J.; Yu, J.Q.; Chen, Z. The perplexing role of autophagy in plant innate immune responses. Mol. Plant Pathol. 2014, 15, 637–645, doi:10.1111/mpp.12118.
- Lai, Z.; Wang, F.; Zheng, Z.; Fan, B.; Chen, Z. A critical role of autophagy in plant resistance to necrotrophic fungal pathogens. Plant J. 2011, 66, 953–968, doi:10.1111/j.1365-313X.2011.04553.x.
- Derrien, B.; Baumberger, N.; Schepetilnikov, M.; Viotti, C.; De Cillia, J.; Ziegler-Graff, V.; Isono, E.; Schumacher, K.; Genschik, P. Degradation of the antiviral component ARGONAUTE1 by the autophagy pathway. Proc. Natl. Acad. Sci. USA 2012, 109, 15942–15946, doi:10.1073/pnas.1209487109.
- Hafren, A.; Macia, J.L.; Love, A.J.; Milner, J.J.; Drucker, M.; Hofius, D. Selective autophagy limits cauliflower mosaic virus infection by NBR1-mediated targeting of viral capsid protein and particles. Proc. Nalt. Acad. Sci. USA 2017, 114, E2026–E2035, doi:10.1073/pnas.1610687114.
- Hafren, A.; Ustun, S.; Hochmuth, A.; Svenning, S.; Johansen, T.; Hofius, D. Turnip Mosaic Virus Counteracts Selective Autophagy of the Viral Silencing Suppressor HCpro. Plant Physiol 2018, 176, 649–662, doi:10.1104/pp.17.01198.
- Nakahara, K.S.; Masuta, C.; Yamada, S.; Shimura, H.; Kashihara, Y.; Wada, T.S.; Meguro, A.; Goto, K.; Tadamura, K.; Sueda, K.; et al. Tobacco calmodulin-like protein provides secondary defense by binding to and directing degradation of virus RNA silencing suppressors. Proc. Natl. Acad. Sci. USA 2012, 109, 10113–10118, doi:10.1073/pnas.1201628109.
- Bassham, D.C.; Laporte, M.; Marty, F.; Moriyasu, Y.; Ohsumi, Y.; Olsen, L.J.; Yoshimoto, K. Autophagy in development and stress responses of plants. Autophagy 2006, 2, 2–11, doi:10.4161/auto.2092.
- Hanamata, S.; Kurusu, T.; Kuchitsu, K. Roles of autophagy in male reproductive development in plants. Front. Plant Sci. 2014, 5, 457, doi:10.3389/fpls.2014.00457.
- Harrison-Lowe, N.J.; Olsen, L.J. Autophagy protein 6 (ATG6) is required for pollen germination in Arabidopsis thaliana. Autophagy 2008, 4, 339–348, doi:10.4161/auto.5629.
- Kurusu, T.; Koyano, T.; Hanamata, S.; Kubo, T.; Noguchi, Y.; Yagi, C.; Nagata, N.; Yamamoto, T.; Ohnishi, T.; Okazaki, Y.; et al. OsATG7 is required for autophagy-dependent lipid metabolism in rice postmeiotic anther development. Autophagy 2014, 10, 878–888, doi:10.4161/auto.28279.
- Michaeli, S.; Galili, G.; Genschik, P.; Fernie, A.R.; Avin-Wittenberg, T. Autophagy in Plants--What's New on the Menu? Trends Plant Sci. 2016, 21, 134–144, doi:10.1016/j.tplants.2015.10.008.
- Stephani, M.; Dagdas, Y. Plant Selective Autophagy-Still an Uncharted Territory With a Lot of Hidden Gems. J. Mol. Biol. 2020, 432, 63–79, doi:10.1016/j.jmb.2019.06.028.
- Bu, F.; Yang, M.K.; Guo, X.; Huang, W.; Chen, L. Multiple Functions of ATG8 Family Proteins in Plant Autophagy. Front. Cell Dev. Biol. 2020, 8, doi:10.3389/fcell.2020.00466.
- Ran, J.; Hashimi, S.M.; Liu, J.Z. Emerging Roles of the Selective Autophagy in Plant Immunity and Stress Tolerance. Int. J. Mol. Sci. 2020, 21, 6321, doi:10.3390/ijms21176321.
- Svenning, S.; Lamark, T.; Krause, K.; Johansen, T. Plant NBR1 is a selective autophagy substrate and a functional hybrid of the mammalian autophagic adapters NBR1 and p62/SQSTM1. Autophagy 2011, 7, 993–1010.
- Avin-Wittenberg, T.; Michaeli, S.; Honig, A.; Galili, G. ATI1, a newly identified atg8-interacting protein, binds two different Atg8 homologs. Plant Signal. Behav. 2012, 7, 685–687, doi:10.4161/psb.20030.
- Honig, A.; Avin-Wittenberg, T.; Ufaz, S.; Galili, G. A New Type of Compartment, Defined by Plant-Specific Atg8-Interacting Proteins, Is Induced upon Exposure of Arabidopsis Plants to Carbon Starvation. Plant Cell 2012, 24, 288–303, doi:10.1105/tpc.111.093112.
- Michaeli, S.; Honig, A.; Levanony, H.; Peled-Zehavi, H.; Galili, G. Arabidopsis ATG8-INTERACTING PROTEIN1 is involved in autophagy-dependent vesicular trafficking of plastid proteins to the vacuole. Plant Cell 2014, 26, 4084–4101, doi:10.1105/tpc.114.129999.
- Zhou, J.; Wang, Z.; Wang, X.; Li, X.; Zhang, Z.; Fan, B.; Zhu, C.; Chen, Z. Dicot-specific ATG8-interacting ATI3 proteins interact with conserved UBAC2 proteins and play critical roles in plant stress responses. Autophagy 2018, 1–18, doi:10.1080/15548627.2017.1422856.
- Hu, S.; Ye, H.; Cui, Y.; Jiang, L.W. AtSec62 is critical for plant development and is involved in ER-phagy in Arabidopsis thaliana. J. Integr. Plant Biol. 2020, 62, 181–200, doi:10.1111/jipb.12872.
- Zhang, X.G.; Ding, X.X.; Marshall, R.S.; Paez-Valencia, J.; Lacey, P.; Vierstra, R.D.; Otegui, M.S. Reticulon proteins modulate autophagy of the endoplasmic reticulum in maize endosperm. Elife 2020, 9, doi:10.7554/eLife.51918.
- Stephani, M.; Picchianti, L.; Gajic, A.; Beveridge, R.; Skarwan, E.; Hernandez, V.S.D.; Mohseni, A.; Clavel, M.; Zeng, Y.L.; Naumann, C.; et al. A cross-kingdom conserved ER-phagy receptor maintains endoplasmic reticulum homeostasis during stress. Elife 2020, 9, doi:10.7554/eLife.58396.
- Vanhee, C.; Zapotoczny, G.; Masquelier, D.; Ghislain, M.; Batoko, H. The Arabidopsis multistress regulator TSPO is a heme binding membrane protein and a potential scavenger of porphyrins via an autophagy-dependent degradation mechanism. Plant Cell 2011, 23, 785–805, doi:10.1105/tpc.110.081570.
- Li, X.; Liu, Q.; Feng, H.; Deng, J.; Zhang, R.; Wen, J.; Dong, J.; Wang, T. Dehydrin MtCAS31 promotes autophagic degradation under drought stress. Autophagy 2020, 16, 862–877, doi:10.1080/15548627.2019.1643656.
- Yang, F.; Kimberlin, A.N.; Elowsky, C.G.; Liu, Y.F.; Gonzalez-Solis, A.; Cahoon, E.B.; Alfano, J.R. A Plant Immune Receptor Degraded by Selective Autophagy. Mol. Plant 2019, 12, 113–123, doi:10.1016/j.molp.2018.11.011.
- Devarenne, T.P. The plant cell death suppressor Adi3 interacts with the autophagic protein Atg8h. Biochem. Bioph. Res. Co. 2011, 412, 699–703, doi:10.1016/j.bbrc.2011.08.031.
- Nolan, T.M.; Brennan, B.; Yang, M.; Chen, J.; Zhang, M.; Li, Z.; Wang, X.; Bassham, D.C.; Walley, J.; Yin, Y. Selective Autophagy of BES1 Mediated by DSK2 Balances Plant Growth and Survival. Dev. Cell 2017, 41, 33–46 e37, doi:10.1016/j.devcel.2017.03.013.
- Zhan, N.; Wang, C.; Chen, L.C.; Yang, H.J.; Feng, J.; Gong, X.Q.; Ren, B.; Wu, R.; Mu, J.Y.; Li, Y.S.; et al. S-Nitrosylation Targets GSNO Reductase for Selective Autophagy during Hypoxia Responses in Plants. Mol. Cell 2018, 71, 142–154, doi:10.1016/j.molcel.2018.05.024.
- Johansen, T.; Lamark, T. Selective autophagy mediated by autophagic adapter proteins. Autophagy 2011, 7, 279–296.
- Zientara-Rytter, K.; Lukomska, J.; Moniuszko, G.; Gwozdecki, R.; Surowiecki, P.; Lewandowska, M.; Liszewska, F.; Wawrzynska, A.; Sirko, A. Identification and functional analysis of Joka2, a tobacco member of the family of selective autophagy cargo receptors. Autophagy 2011, 7, 1145–1158, doi:10.4161/auto.7.10.16617.
- Lamark, T.; Kirkin, V.; Dikic, I.; Johansen, T. NBR1 and p62 as cargo receptors for selective autophagy of ubiquitinated targets. Cell Cycle 2009, 8, 1986–1990, doi:10.4161/cc.8.13.8892.
- Young, P.G.; Passalacqua, M.J.; Chappell, K.; Llinas, R.J.; Bartel, B. A facile forward-genetic screen for Arabidopsis autophagy mutants reveals twenty-one loss-of-function mutations disrupting six ATG genes. Autophagy 2019, 15, 941–959, doi:10.1080/15548627.2019.1569915.
- Zhou, J.; Zhang, Y.; Qi, J.; Chi, Y.; Fan, B.; Yu, J.Q.; Chen, Z. E3 Ubiquitin Ligase CHIP and NBR1-Mediated Selective Autophagy Protect Additively against Proteotoxicity in Plant Stress Responses. PLoS Genet. 2014, 10, e1004116, doi:10.1371/journal.pgen.1004116.
- Jung, H.; Lee, H.N.; Marshall, R.S.; Lomax, A.W.; Yoon, M.J.; Kim, J.; Kim, J.H.; Vierstra, R.D.; Chung, T. Arabidopsis cargo receptor NBR1 mediates selective autophagy of defective proteins. J. Exp. Bot. 2020, 71, 73–89, doi:10.1093/jxb/erz404.
- Ji, C.Y.; Zhou, J.; Guo, R.F.; Lin, Y.S.; Kung, C.H.; Hu, S.; Ng, W.Y.; Zhuang, X.H.; Jiang, L.W. AtNBR1 Is a Selective Autophagic Receptor for AtExo70E2 in Arabidopsis. Plant Physiol. 2020, 184, 777–791, doi:10.1104/pp.20.00470.
- Ustun, S.; Hofius, D. Anti- and pro-microbial roles of autophagy in plant-bacteria interactions. Autophagy 2018, 14, 1465–1466, doi:10.1080/15548627.2018.1475817.
- Sedaghatmehr, M.; Thirumalaikumar, V.P.; Kamranfar, I.; Marmagne, A.; Masclaux-Daubresse, C.; Balazadeh, S. A regulatory role of autophagy for resetting the memory of heat stress in plants. Plant Cell Environ. 2019, 42, 1054–1064, doi:10.1111/pce.13426.
- Thirumalaikumar, V.P.; Gorka, M.; Schulz, K.; Masclaux-Daubresse, C.; Sampathkumar, A.; Skirycz, A.; Vierstra, R.D.; Balazadeh, S. Selective autophagy regulates heat stress memory in Arabidopsis by NBR1-mediated targeting of HSP90 and ROF1. Autophagy 2020, 1–16, doi:10.1080/15548627.2020.1820778.
- Meiri, D.; Breiman, A. Arabidopsis ROF1 (FKBP62) modulates thermotolerance by interacting with HSP90.1 and affecting the accumulation of HsfA2-regulated sHSPs. Plant J. 2009, 59, 387–399, doi:10.1111/j.1365-313X.2009.03878.x.
- Liu, J.; Guo, W. The exocyst complex in exocytosis and cell migration. Protoplasma 2012, 249, 587–597, doi:10.1007/s00709-011-0330-1.
- Ding, Y.; Wang, J.; Chun Lai, J.H.; Ling Chan, V.H.; Wang, X.; Cai, Y.; Tan, X.; Bao, Y.; Xia, J.; Robinson, D.G.; et al. Exo70E2 is essential for exocyst subunit recruitment and EXPO formation in both plants and animals. Mol. Biol. Cell 2014, 25, 412–426, doi:10.1091/mbc.E13-10-0586.
- Wang, J.; Ding, Y.; Wang, J.; Hillmer, S.; Miao, Y.; Lo, S.W.; Wang, X.; Robinson, D.G.; Jiang, L. EXPO, an exocyst-positive organelle distinct from multivesicular endosomes and autophagosomes, mediates cytosol to cell wall exocytosis in Arabidopsis and tobacco cells. Plant Cell 2010, 22, 4009–4030, doi:10.1105/tpc.110.080697.
- Niemiro, A.; Cysewski, D.; Brzywczy, J.; Wawrzynska, A.; Sienko, M.; Poznanski, J.; Sirko, A. Similar but Not Identical-Binding Properties of LSU (Response to Low Sulfur) Proteins FromArabidopsis thaliana. Front. Plant Sci. 2020, 11, doi:10.3389/fpls.2020.01246.
- Tarnowski, L.; Rodriguez, M.C.; Brzywczy, J.; Cysewski, D.; Wawrzynska, A.; Sirko, A. Overexpression of the Selective Autophagy Cargo Receptor NBR1 Modifies Plant Response to Sulfur Deficit. Cells Basel. 2020, 9, 669, doi:10.3390/cells9030669.
- Tarnowski, L.; Rodriguez, M.C.; Brzywczy, J.; Piecho-Kabacik, M.; Krckova, Z.; Martinec, J.; Wawrzynska, A.; Sirko, A. A selective autophagy cargo receptor NBR1 modulates abscisic acid signalling in Arabidopsis thaliana. Sci. Rep. UK 2020, 10, doi:10.1038/s41598-020-64765-z.
