The nanodelivery system plays a dual function in RNA-based vaccination by acting as a carrier system and as an adjuvant. That is due to its similarity to microorganisms structurally and size-wise; the nanodelivery system can augment the response by the immune system via simulating the natural infection process. Nanodelivery systems allow non-invasive mucosal administration, targeted immune cell delivery, and controlled delivery, reducing the need for multiple administrations. They also allow co-encapsulating with immunostimulators to improve the overall adjuvant capacity.
- mRNA
- adjuvant
- vaccine
- nanoparticles
- nanodelivery systems
- lipids
- polymers
1. Introduction
Messenger RNA (mRNA)-based vaccines have shown promise as techniques for the development of vaccines against infectious diseases and several types of cancer since the 2000s. They have been evaluated in multiple studies on the prophylaxis and treatment of infectious diseases, several types of cancer, and autoimmune diseases. Clinical studies have demonstrated their potential in terms of safety and immune responses. As prophylactic vaccines in infectious diseases, clinical trials are still in their early stages, mostly phase I clinical studies, with most trials testing mRNA-based vaccines for coronavirus disease 2019 (COVID-19) and rabies. Other studied conditions include Zika virus and H10N8 and H7N9 influenza viruses. As therapeutic vaccines for cancer immunotherapy, many mRNA-based vaccines have proceeded to phase II clinical trials, including those for melanoma, non-small-cell lung carcinoma, and prostate cancer. Examples of clinical trials and their results are summarized in Table 1.
Table 1. Examples of clinical trials using nanodelivery systems for mRNA-based vaccines.
| Disease | Target Antigen | Route/Nanodelivery System Type | Phase | Status/Results | Reference | |||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
Rabies | Rabies virus glycoprotein | IM; ID; protamine complex | I | (Completed) Produced boostable neutralizing antibodies when administered with a needle-free device (spring-powered ID and IM injectors, carbon dioxide gas-powered ID injector); vaccine appeared to be safe, with no serious adverse effects except for a case of temporary moderate Bell’s palsy |
[2] |
[1] | NCT02241135 | |||||||||||
H10N8 and H7N9 Influenza Viruses | Hemagglutinin glycoprotein from the H10N8 influenza strain or the H7N9 influenza strain | IM; ID; lipid nanoparticles | I | (H10N8 vaccine: Completed; H7N9 vaccine: Active, not recruiting). Vaccines produced a strong humoral immune response in healthy adults with no serious adverse effects |
[3] |
[2] | NCT03345043 | |||||||||||
COVID-19 | SARS-CoV-2 spike (S) glycoprotein | IM; lipid nanoparticles | I | (Active, not recruiting) Provoked high levels of binding and neutralizing antibodies in younger and older adults and the responses were similar those seen in COVID-19-recovered patients; no serious adverse effects were reported | NCT04283461 | |||||||||||||
SARS-CoV-2 spike (S) glycoprotein | IM; lipid nanoparticles | I/II | (Recruiting) No results posted | NCT04480957 | ||||||||||||||
Two vaccines: BNT162b1, encoding a secreted trimerized SARS-CoV-2 receptor-binding domain; BNT162b2, encoding a prefusion stabilized membrane-anchored SARS-CoV-2 full-length spike | IM; lipid nanoparticles | I/II | (Recruiting) Both vaccines stimulated neutralizing antibodies in younger and older adults that are similar or higher than COVID-19-recovered patients; BNT162b2 was associated with less systemic reactions, especially in older participants; no serious adverse effects were reported | NCT04368728 | ||||||||||||||
Prostate Cancer | Prostate-specific membrane antigen, prostate stem cell antigen, and six-transmembrane epithelial antigen of the prostate 1 | ID; protamine complex | I/IIa | (Terminated) Induction of CD4+ and CD8+ T-cell responses; probably vaccine-related urinary retention occurred in 3 patients |
[7] |
[6] | NCT01817738 | |||||||||||
Non-small Cell Lung Cancer | Non-small cell lung cancer antigens: New York esophageal squamous cell carcinoma (NY-ESO-1), melanoma antigen family (MAGE) C1 and C2, baculoviral inhibitor of apoptosis repeat-containing 5, trophoblast glycoprotein, and mucin-1 antigen | ID; protamine complex | Ib | (Terminated) Induction of immune response against the six encoded antigens; no vaccine-related serious adverse effects were reported |
[8] |
[7] | NCT01915524 | |||||||||||
Melanoma | NY-ESO-1, MAGE-A3, tyrosinase and TPTE | Intravenous; lipoplex | I | (Active, not recruiting) Induction of IFN-α and strong antigen-specific T-cell responses |
[9] |
[8] | NCT02410733 |
mRNA: messenger RNA; ID: intradermal; IM: intramuscular; COVID-19: coronavirus disease 2019; SARS-CoV-2: Severe acute respiratory syndrome coronavirus 2; IFN-α: interferons-α.
mRNA-based vaccines work by using the host cell’s translation mechanism to produce the relevant antigen and trigger the adaptive immune response. After entering the cytosol, the cell treats it as an endogenous mRNA and translation starts instantly (Figure 1) [1][9].
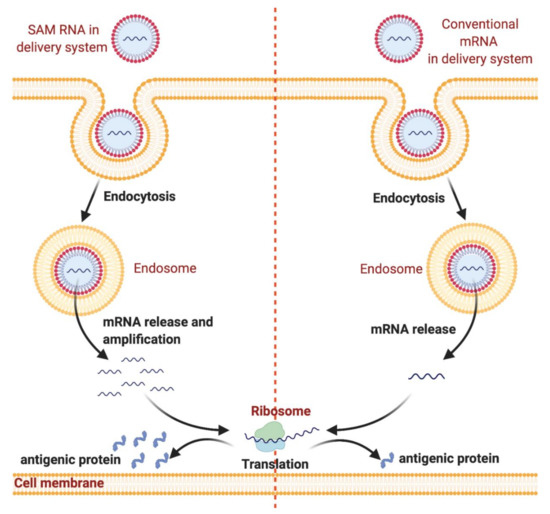
Nanodelivery systems protect messenger RNA (mRNA) from degradation and enhance endocytosis and endosomal escape. mRNA can be loaded into nanodelivery systems for protection from enzymatic degradation. After administration, mRNA is internalized by dendritic cells through endocytosis followed by endosomal escape. Subsequently, mRNA is released into the cytosol, which is followed by mRNA translation into antigenic protein by ribosomes. The figure was created using
.
The immune system can also be triggered when extracellular mRNA is recognized by pattern recognition receptors (PRRs): endosomal membrane-located Toll-like receptors (TLRs 3, 7, and 8) and cytosolic nucleic acid sensors such as retinoic acid-inducible gene (RIG)-I-like receptors (RLRs) (Figure 2). When activated, PRRs provoke type I interferon (IFN) responses, which may be strong and negatively affect antigen expression and may cause T-cell exhaustion. However, the formulation of the vaccine impacts the types of sensors activated. During the in vitro production, mRNA capping is necessary to be recognized as an endogenous molecule. This could be achieved using cap analogues or through enzymatic capping. Double-stranded RNAs (dsRNA) resulting from the transcription reaction should be removed to reduce type I IFN production and enhance antigen expression through high-performance liquid chromatography (HPLC) or cellulose purification [10]. mRNA vaccines can be divided into two types: conventional mRNA-based vaccines and self-amplifying mRNA (SAM RNA) vaccines. Conventional mRNA-based vaccines harbor only the antigen gene of interest and cannot amplify itself, whereas the SAM RNA vaccines each contain an engineered RNA virus genome that comprises the virus’s non-structural protein genes, which are essential in the RNA replication machinery, and the antigen gene of interest. After its introduction into the host cell, a SAM vaccine is able to amplify, resulting in replicons that are able to produce high amounts of the antigen gene. Replicons are unable to produce infectious virions as they lack the structural protein genes, and thus, cannot spread to the neighboring cells [11]. SAM RNA vaccines are designed to enhance the extent of protein expression and the induced immune response. The use of mRNA-based vaccines has multiple advantages over conventional whole-organism, subunit, and DNA-based vaccines, including safety profile, ability to be rapidly developed, low-cost manufacturing potential, and high potency. mRNA-based vaccines are considered to be relatively safe as there is no associated risk of infection as experienced with live attenuated vaccines [12]. Additionally, compared to DNA-based vaccines, there is no need for the mRNA to enter the nucleus; thus, there is no risk of genomic integration and unpredictable long-term expression is avoided, allowing control of the treatment duration and side effects [13]. Moreover, mRNA production is cell-free, using in vitro transcription (IVT) methods that highly decrease bacterial contamination and permit rapid scale-up over a short period and low-cost manufacturing due to the high yields [1][9]. A new purification method using cellulose have been developed recently which helps in further reducing the manufacturing costs. In comparison to HPLC, this method does not require high-cost equipment, fast, with similar antigen expression and more mRNA recovery rate in comparison to mRNA--based vaccines purified by HPLC [14]. Thus, mRNA-based vaccines are good candidates for responding to the COVID-19 pandemic. The design and manufacturing of mRNA-based vaccines on the clinical scale is possible within weeks when the viral antigen sequence becomes available. For instance, only 42 days were required for Moderna’s mRNA-1273 (Cambridge, MA, USA) to enter phase I clinical testing after the sequencing of the full SARS-CoV-2 genome (ClinicalTrials.gov identifier NCT04283461). Recently, mRNA-1273 has entered into phase III clinical studies (ClinicalTrials.gov identifier NCT04470427).
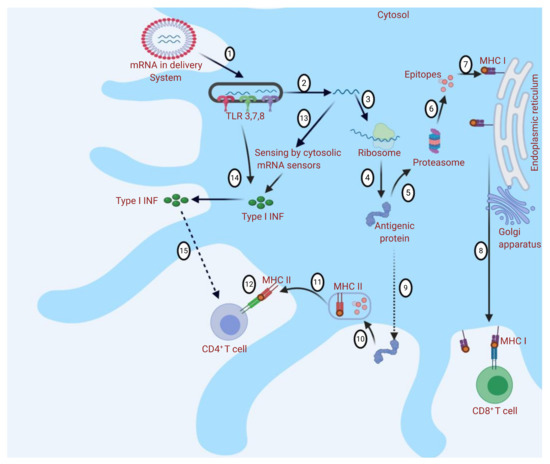
Different immune responses to messenger RNA (mRNA)-based vaccine nanodelivery systems: The mRNA delivery system is engulfed into the cell by endocytosis, and Toll-like receptors (TLRs) are activated (1). mRNA is released into the cytosol (2). mRNA binds to ribosome and translation occurs (3 and 4). Antigenic protein provokes the immune response through major histocompatibility complex (MHC I or MHC II) presentation (5 and 9). (5) In MHC I presentation, peptides are produced by proteolysis (6), and CD8+ T cells are stimulated (7 and 8). In MHC II presentation (9), peptides are produced by endosomal proteolysis (10 and 11), and CD4+ T cells are stimulated (12). Cytosolic sensing of intracellular mRNA leads to activation of retinoic acid-inducible gene (RIG)-I-like receptors (RLRs) and nucleotide-binding oligomerization domain (NOD)-like receptors (NLRs). Activation of TLRs, RLRs, and NLRs induces the production of type I interferons (IFNs) (13 and 14). Type I IFNs may have a positive or negative effect on the activation of T cells (as they could determine the differentiation of antigen-primed CD8
T cells into cytotoxic effectors but may also cause T-cell exhaustion) (15). The figure was created using
.
Until recently, mRNA vaccine applications have been restricted by instability and inefficient in vivo delivery of mRNA molecules. Their delivery to the cytosol is hindered by the rapid degradation of naked mRNA by ribonuclease and the negatively charged and high-molecular-weight (105–106 Da) mRNA molecule, preventing passive diffusion across the cell membrane [15,16][15][16]. Although local intranodal injection of RNA leads to the effective activation of a specific immune response [17[17][18][19],18,19], delivery systems are crucial for successful in vivo delivery of mRNA to the site of action and for large-scale preventive vaccinations.
Ideally, the in vivo delivery system should provide protection against degradation of the mRNA molecule by extracellular RNAses, facilitate cellular uptake, and target antigen-presenting cells (APCs). Multiple methods have been studied to enhance mRNA delivery. These include physical methods, viral-based vectors, and non-viral vectors. Physical methods include using a gene gun and electroporation; both methods have been tested and shown to improve delivery to cells [20,21][20][21]. However, physical methods often destroy the cells, making them unsuitable for mRNA delivery [22,23,24][22][23][24]. Viral-based vectors have also been studied as mRNA delivery systems, wherein the genes of adeno-associated viruses are replaced completely or partially with the gene of interest. A drawback of this method is the risk of genomic integration [25,26,27][25][26][27]. Non-viral victors include nanodelivery systems (Figure 3): lipid-based, polymer-based, and lipid–polymer hybrid nanoparticles [28,29,30,31,32,33,34][28][29][30][31][32][33][34]. These are currently the most favored methods for delivering mRNA, as they are considered safe, stable, and low-cost, and provide highly efficient transfection. They provide the advantage of acting as delivery systems and as adjuvants. Due to their resemblance to pathogens in size and structure, nanoparticles can induce immunogenicity by mimicking the natural infection activity. Nanodelivery systems also allow non-invasive mucosal administration, targeted immune cell delivery, and controlled delivery, reducing the need for multiple administrations. Moreover, they allow co-encapsulating with immunostimulators to improve the overall adjuvant capacity [27]. Furthermore, it was reported that when nanoparticles (of 10–100 nm size) injected into tissue (i.e., through intramuscular, intradermal, subcutaneous or intraperitoneal) they are readily carried across the lymphatic endothelium by interstitial fluid, but they are too big to diffuse into the blood circulatory system. These nanoparticles are found to have enhanced lymph node transport efficiencies which is very important for vaccine developments [34]. Because improved lymphatic transport permits direct access to dendritic cells in lymphoid node and consequently facilitates the antigen uptake and presentation to B cells enhancing the antibody-mediated immunity [35], or to T cells, allowing cell-mediated immunity [36,37][36][37]. More details on the importance of the nanodelivery systems in the vaccines development have been described in recent reviews [38,39][38][39].
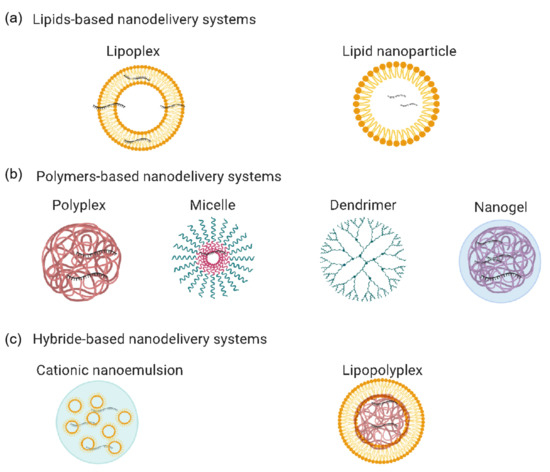
Figure 3. mRNA-based vaccine nanodelivery systems: (a) lipid-based nanodelivery systems, including lipoplexes and lipid nanoparticles; (b) polymer-based nanodelivery systems, including polyplexes, micelles, dendrimers, and nanogels; (c) hybrid-based nanodelivery systems, including cationic nanoemulsions and lipopolyplexes. The figure was generated using Biorender.com.
2. mRNA-Based Vaccines’ Nanodelivery Systems
2.1. Lipid-Based Nanodelivery Systems
Currently, lipid nanoparticles and lipoplexes are among the most commonly used lipid-based nanodelivery systems for in vivo delivery of mRNA molecules [29]. The general chemical structure of a cationic lipid consists of a hydrophobic chain attached to a head group. The chain is a hydrophobic chain that is symmetric or di-symmetric, modified or unmodified, saturated or unsaturated, and linear or branched. The cationic head group holds a linker molecule that can be cleavable to enhance biodegradation. The head group usually holds at least one group, for instance, amine, which ionizes and changes its negative charge to a positive charge at physiological pH. Accordingly, cationic lipids can be complexed with anionic molecules via electrostatic interactions [47][40].
2.2. Polymer-Based Nanodelivery Systems
Diethylaminoethyl (DEAE) dextran is a polycationic derivative of the carbohydrate polymer dextran. It was the first cationic polymer to be examined as a delivery system for mRNA [104][41]. Later, it was demonstrated that mRNA transfection through lipid-based delivery systems is 100 to 1000 times more efficient than DEAE–dextran-based delivery systems [105][42]. This finding slowed the advancement of polymer-based delivery systems and promoted the progress of lipid-based delivery systems for mRNA and other nucleic acids. However, cationic polymers can be considered suitable partners for noncovalent interactions with nucleic acids, which resulted in satisfactory in vivo transfection. Cationic polymers provide considerable flexibility in terms of structure modifications and development. Some sequence-defined polymers are advantageous for demonstrating fine structure–activity relationships. Hence, cationic polymers have attracted substantial interest as non-viral delivery systems in the area of nucleic acid delivery. In the last few years, many cationic polymers have been developed, examined, and used as efficient delivery systems for nucleic acids [106][43]. However, applications of cationic polymers as nanodelivery systems for mRNA have not been thoroughly explored compared to pDNA and siRNA; they have the potential to compete with many well-studied lipid-based delivery systems [107][44].
2.3. Hybrid-Based Nanodelivery Systems
The nanodelivery systems for mRNA vaccines may consist of many chemical compounds, including lipids, polymers, and peptides in one structure for more potent transfection. These nanodelivery systems can be classified as hybrid nanodelivery systems. These hybrid nanodelivery systems normally integrate the potential advantages of their constituents and offer more flexibility in comparison with non-hybrid nanodelivery systems [27,32][27][32].
References
- Alberer, M.; Gnad-Vogt, U.; Hong, H.S.; Mehr, K.T.; Backert, L.; Finak, G.; Gottardo, R.; Bica, M.A.; Garofano, A.; Koch, S.D.; et al. Safety and immunogenicity of a mRNA rabies vaccine in healthy adults: An open-label, non-randomised, prospec-tive, first-in-human phase 1 clinical trial. Lancet 2017, 390, 1511–1520, doi:10.1016/s0140-6736(17)31665-3.
- Feldman, R.A.; Fuhr, R.; Smolenov, I.; Ribeiro, A. (Mick); Panther, L.; Watson, M.; Senn, J.J.; Smith, M.; AlmarssonӦ.; Pujar, H.S.; et al. mRNA vaccines against H10N8 and H7N9 influenza viruses of pandemic potential are immunogenic and well tolerated in healthy adults in phase 1 randomized clinical trials. Vaccine 2019, 37, 3326–3334, doi:10.1016/j.vaccine.2019.04.074.
- Jackson, L.A.; Anderson, E.J.; Rouphael, N.G.; Roberts, P.C.; Makhene, M.; Coler, R.N.; McCullough, M.P.; Chappell, J.D.; Denison, M.R.; Stevens, L.J.; et al. An mRNA Vaccine against SARS-CoV-2—Preliminary Report. N. Engl. J. Med. 2020, 383, 1920–1931, doi:10.1056/nejmoa2022483.
- Anderson, E.J.; Rouphael, N.G.; Widge, A.T.; Jackson, L.A.; Roberts, P.C.; Makhene, M.; Chappell, J.D.; Denison, M.R.; Ste-vens, L.J.; Pruijssers, A.J.; et al. Safety and Immunogenicity of SARS-CoV-2 mRNA-1273 Vaccine in Older Adults. N. Engl. J. Med. 2020 383, 2427–2438, doi:10.1056/nejmoa2028436.
- Mulligan, M.J.; Lyke, K.E.; Kitchin, N.; Absalon, J.; Gurtman, A.; Lockhart, S.P.; Li, P. Phase 1/2 Study to Describe the Safety and Immunogenicity of a COVID-19 RNA Vaccine Candidate (BNT162b1) in Adults 18 to 55 Years of Age: Interim Report. medRxiv 2020. https://doi.org/10.1101/2020.06.30.20142570.
- Walsh, E.E.; Frenck, R.; Falsey, A.R.; Kitchin, N.; Absalon, J.; Gurtman, A.; Swanson, K.A. RNA-Based COVID-19 Vaccine BNT162b2 Selected for a Pivotal Efficacy Study. Prepr. Serv. Heal. Sci. 2020, doi:10.1101/2020.08.17.20176651.
- Kübler, H.R.; Scheel, B.; Gnad-Vogt, U.; Miller, K.; Schultze-Seemann, W.; Dorp, F.V.; Parmiani, G.; Hampel, C.; Wedel, S.; Trojan, L.; et al. Self-adjuvanted mRNA vaccination in advanced prostate cancer patients: a first-in-man phase I/IIa study. J. Immunother. Cancer 2015, 3, 26, doi:10.1186/s40425-015-0068-y.
- Papachristofilou, A.; Hipp, M.M.; Klinkhardt, U.; Früh, M.; Sebastian, M.; Weiss, C.; Pless, P.D.D.M.; Cathomas, R.; Hilbe, W.; Pall, G.; et al. Phase Ib evaluation of a self-adjuvanted protamine formulated mRNA-based active cancer immunothera-py, BI1361849 (CV9202), combined with local radiation treatment in patients with stage IV non-small cell lung cancer. J. Im-munother. Cancer 2019, 7, 38, doi:10.1186/s40425-019-0520-5.
- Iavarone, C.; O’Hagan, D.T.; Yu, D.; Delahaye, N.F.; Ulmer, J.B. Mechanism of action of mRNA-based vaccines. Expert Rev. Vaccines 2017, 16, 871–881, doi:10.1080/14760584.2017.1355245.
- Chandler, M.; Johnson, M.B.; Panigaj, M.; A Afonin, K. Innate immune responses triggered by nucleic acids inspire the design of immunomodulatory nucleic acid nanoparticles (NANPs). Curr. Opin. Biotechnol. 2020, 63, 8–15, doi:10.1016/j.copbio.2019.10.011.
- Kariko, K.; Muramatsu, H.; Welsh, F.A.; Ludwig, J.; Kato, H.; Akira, S.; Weissman, D. Incorporation of Pseudouridine into mRNA Yields Superior Nonimmunogenic Vector with Increased Translational Capacity and Biological Stability. Mol. Ther. 2008, 16, 1833–1840, doi:10.1038/mt.2008.200.
- Jahanafrooz, Z.; Baradaran, B.; Mosafer, J.; Hashemzaei, M.; Rezaei, T.; Mokhtarzadeh, A.; Hamblin, M.R. Comparison of DNA and mRNA vaccines against cancer. Drug Discov. Today 2020, 25, 552–560, doi:10.1016/j.drudis.2019.12.003.
- Baiersdörfer, M.; Boros, G.; Muramatsu, H.; Mahiny, A.; Vlatkovic, I.; Sahin, U.; Karikó, K. A Facile Method for the Removal of dsRNA Contaminant from In Vitro-Transcribed mRNA. Mol. Ther. Nucl. Acids 2019, 15, 26–35, doi:10.1016/j.omtn.2019.02.018.
- Kowalski, P.S.; Rudra, A.; Miao, L.; Anderson, D.G. Delivering the Messenger: Advances in Technologies for Therapeutic mRNA Delivery. Mol. Ther. 2019, 27, 710–728, doi:10.1016/j.ymthe.2019.02.012.
- Sharova, L.V.; Sharov, A.A.; Nedorezov, T.; Piao, Y.; Shaik, N.; Ko, M.S. Database for mRNA Half-Life of 19 977 Genes Ob-tained by DNA Microarray Analysis of Pluripotent and Differentiating Mouse Embryonic Stem Cells. DNA Res. 2008, 16, 45–58, doi:10.1093/dnares/dsn030.
- Kreiter, S.; Selmi, A.; Diken, M.; Koslowski, M.; Britten, C.M.; Huber, C.; Türeci, Özlem; Sahin, U. Intranodal Vaccination with Naked Antigen-Encoding RNA Elicits Potent Prophylactic and Therapeutic Antitumoral Immunity. Cancer Res. 2010, 70, 9031–9040, doi:10.1158/0008-5472.can-10-0699.
- Diken, M.; Kreiter, S.; Selmi, A.; Britten, C.M.; Huber, C.; Tureci, O.; Sahin, U. Selective uptake of naked vaccine RNA by dendritic cells is driven by macropinocytosis and abrogated upon DC maturation. Gene Ther. 2011, 18, 702–708, doi:10.1038/gt.2011.17.
- van Lint, S.; Goyvaerts, C.; Maenhout, S.; Goethals, L.; Disy, A.; Benteyn, D.; Pen, J.; Bonehill, A.; Heirman, C.; Breckpot, K.; et al. Preclinical Evaluation of TriMix and Antigen mRNA-Based Antitumor Therapy. Cancer Res. 2012, 72, 1661–1671, doi:10.1158/0008-5472.can-11-2957.
- van Tendeloo, V.; Snoeck, H.-W.; Lardon, F.; Vanham, G.; Nijs, G.; Lenjou, M.; Hendriks, L.; van Broeckhoven, C.; Moulijn, A.; Rodrigus, I.; et al. Nonviral transfection of distinct types of human dendritic cells: High-efficiency gene transfer by elec-troporation into hematopoietic progenitor- but not monocyte-derived dendritic cells. Gene Ther. 1998, 5, 700–707, doi:10.1038/sj.gt.3300626.
- Qiu, P.; Ziegelhoffer, P.; Sun, J.; Yang, N.S. Gene gun delivery of mRNA in situ results in efficient transgene expression and genetic immunization. Gene Ther. 1996, 3.
- de Temmerman, M.-L.; Dewitte, H.; Vandenbroucke, R.E.; Lucas, B.; Libert, C.; Demeester, J.; de Smedt, S.C.; Lentacker, I.; Rejman, J. mRNA-Lipoplex loaded microbubble contrast agents for ultrasound-assisted transfection of dendritic cells. Bio-materials 2011, 32, 9128–9135, doi:10.1016/j.biomaterials.2011.08.024.
- van Meirvenne, S.; Straetman, L.; Heirman, C.; Dullaers, M.; de Greef, C.; van Tendeloo, V.; Thielemans, K. Efficient genetic modification of murine dendritic cells by electroporation with mRNA. Cancer Gene Ther. 2002, 9, 787–797, doi:10.1038/sj.cgt.7700499.
- Dewitte, H.; van Lint, S.; Heirman, C.; Thielemans, K.; de Smedt, S.C.; Breckpot, K.; Lentacker, I. The potential of antigen and TriMix sonoporation using mRNA-loaded microbubbles for ultrasound-triggered cancer immunotherapy. J. Control. Re-lease 2014, 194, 28–36, doi:10.1016/j.jconrel.2014.08.011.
- Agapov, E.V.; Frolov, I.; Lindenbach, B.D.; Prágai, B.M.; Schlesinger, S.; Rice, C.M. Noncytopathic Sindbis virus RNA vectors for heterologous gene expression. Proc. Natl. Acad. Sci. USA 1998, 95, 12989–12994, doi:10.1073/pnas.95.22.12989.
- Ferrari, S.; Griesenbach, U.; Shiraki-Iida, T.; Shu, T.; Hironaka, T.; Hou, X.; Williams, J.; Zhu, J.; Jeffery, P.K.; Geddes, D.M.; et al. A defective nontransmissible recombinant Sendai virus mediates efficient gene transfer to airway epithelium in vivo. Gene Ther. 2004, 11, 1659–1664, doi:10.1038/sj.gt.3302334.
- Bitzer, M.; Armeanu, S.; Lauer, U.M.; Neubert, W.J. Sendai virus vectors as an emerging negative-strand RNA viral vector system. J. Gene Med. 2003, 5, 543–553, doi:10.1002/jgm.426.
- Wadhwa, A.; Aljabbari, A.; Lokras, A.; Foged, C.; Thakur, A. Opportunities and Challenges in the Delivery of mRNA-Based Vaccines. Pharmaceutics 2020, 12, 102, doi:10.3390/pharmaceutics12020102.
- Uchida, S.; Perche, F.; Pichon, C.; Cabral, H. Nanomedicine-Based Approaches for mRNA Delivery. Mol. Pharm. 2020, 17, 3654–3684, doi:10.1021/acs.molpharmaceut.0c00618.
- Weng, Y.; Li, C.; Yang, T.; Hu, B.; Zhang, M.; Guo, S.; Xiao, H.; Liang, X.-J.; Huang, Y. The challenge and prospect of mRNA therapeutics landscape. Biotechnol. Adv. 2020, 40, 107534, doi:10.1016/j.biotechadv.2020.107534.
- Maruggi, G.; Zhang, C.; Li, J.; Ulmer, J.B.; Yu, D. mRNA as a Transformative Technology for Vaccine Development to Con-trol Infectious Diseases. Mol. Ther. 2019, 27, 757–772, doi:10.1016/j.ymthe.2019.01.020.
- Zhang, C.; Maruggi, G.; Shan, H.; Li, J. Advances in mRNA Vaccines for Infectious Diseases. Front. Immunol. 2019, 10, 594, doi:10.3389/fimmu.2019.00594.
- Tan, L.; Sun, X. Recent advances in mRNA vaccine delivery. Nano Res. 2018, 11, 5338–5354, doi:10.1007/s12274-018-2091-z.
- Pardi, N.; Hogan, M.J.; Porter, F.W.; Weissman, D. mRNA vaccines—A new era in vaccinology. Nat. Rev. Drug Discov. 2018, 17, 261–279, doi:10.1038/nrd.2017.243.
- Bachmann, M.F.; Jennings, G.T. Vaccine delivery: A matter of size, geometry, kinetics and molecular patterns. Nat. Rev. Im-munol. 2010, 10, 787–796, doi:10.1038/nri2868.
- Reddy, S.T.; van der Vlies, J.; Simeoni, E.; Angeli, V.; Randolph, G.J.; O’Neil, C.P.; Lee, L.K. Exploiting lymphatic transport and complement activation in nanoparticle vaccines. Nat. Biotechnol. 2007, 25, 1159–1164, doi:10.1038/nbt1332.
- Clemente-Casares, X.; Blanco, J.; Ambalavanan, P.; Yamanouchi, J.; Singha, S.; Fandos, C.; Tsai, S.; Wang, J.; Garabatos, N.; Izquierdo, C.; et al. Expanding antigen-specific regulatory networks to treat autoimmunity. Nat. Cell Biol. 2016, 530, 434–440, doi:10.1038/nature16962.
- Singha, S.; Shao, K.; Yang, Y.; Clemente-Casares, X.; Solé, P.; Clemente, A.; Blanco, J.; Dai, Q.; Song, F.; Liu, S.W.; et al. Pep-tide–MHC-based nanomedicines for autoimmunity function as T-cell receptor microclustering devices. Nat. Nanotechnol. 2017, 12, 701–710, doi:10.1038/nnano.2017.56.
- Lung, P.; Yang, J.; Li, Q. Nanoparticle formulated vaccines: Opportunities and challenges. Nanoscale 2020, 12, 5746–5763, doi:10.1039/c9nr08958f.
- Jin, Z.; Gao, S.; Cui, X.; Sun, D.; Zhao, K. Adjuvants and delivery systems based on polymeric nanoparticles for mucosal vac-cines. Int. J. Pharm. 2019, 572, 118731, doi:10.1016/j.ijpharm.2019.118731.
- Zhuang, X.; Qi, Y.; Wang, M.; Yu, N.; Nan, F.; Zhang, H.; Tian, M.; Li, C.; Lu, H.; Jin, N. mRNA Vaccines Encoding the HA Protein of Influenza a H1N1 Virus Delivered by Cationic Lipid Nanoparticles Induce Protective Immune Responses in Mice. Vaccines 2020, 8, 123, doi:10.3390/vaccines8010123.
- Koch, G. Interaction of Poliovirus-Specific RNAs with HeLa Cells and E. coli. Electrodepos. Surface Finish. 1973, 62, 89–138, doi:10.1007/978-3-642-65772-6_4.
- Malone, R.W.; Felgner, P.L.; Verma, I.M. Cationic liposome-mediated RNA transfection. Proc. Natl. Acad. Sci. USA 1989, 86, 6077–6081, doi:10.1073/pnas.86.16.6077.
- Lächelt, U.; Wagner, E. Nucleic Acid Therapeutics Using Polyplexes: A Journey of 50 Years (and Beyond). Chem. Rev. 2015, 115, 11043–11078, doi:10.1021/cr5006793.
- Wang, W.; Li, W.; Ma, N.; Steinhoff, G. Non-Viral Gene Delivery Methods. Curr. Pharm. Biotechnol. 2013, 14, 46–60, doi:10.2174/1389201011314010008.
