In eukaryotes, RNA trans-splicing is a significant RNA modification process for the end-to-end li-gation of exons from separately transcribed primary transcripts to generate mature mRNA. So far, three different categories of RNA trans-splicing have been found in organisms within a diverse range.
- group II intron
- trans-splicing
- ribonucleoprotein complex
- chloroplast
- Chlamydomonas reinhardtii
Note: The following contents are extract from your paper. The entry will be online only after author check and submit it.
1. Introduction
One of the unexpected and outstanding discoveries in 20th century biology was the identification of discontinuous eukaryotic genes [1][2][1,2]. Walter Gilbert introduced the terms “exon” for expressed sequences and “intron” for intervening sequences [3]. In order to fuse exons and generate mature messenger RNA (mRNA), introns are posttranscriptionally removed by splicing. Several studies revealed that splicing in nuclei of eukaryotes is mediated by highly dynamic ribonucleoprotein machinery called the spliceosome [4]. The intervening sequences in the nuclear precursor mRNA of eukaryotes are therefore described as spliceosomal introns or nuclear mRNA introns. Further intron types have been identified and are classified by their structural characteristics and splicing mechanisms, i.e., archaeal introns, group I introns, and group II introns. Spliceosomal introns and group II introns share a common splicing mechanism (as outlined later) that is distinctly different from the splicing mechanism of group I introns, which are found in genomes of bacteria and organelles as well as in nuclei of lower eukaryotes. Finally, archaeal introns are comparably small and share a pre-tRNA intron excision mechanism with eukaryotes.
Here, we summarize our current knowledge about the relationship between group II introns and spliceosomal introns. This includes a brief description of introns-early and introns-late theories, a comparison of sequence homologies, and discussion of mechanistic similarities. We describe nuclear and organellar splicing factors further and discuss how trans-spliced and degenerated group II introns may have evolved into spliceosomal introns.
In the second part of our review, we briefly discuss trans-spliced introns in different organismal lineages and finally summarize the extensive experimental data supporting the existence of a trans-splicing supercomplex in C. reinhardtii chloroplasts.
2. Introns-Early or Introns-Late Debate
Very soon after the discovery of spliceosomal introns, several theories arose about their origin and evolution. The introns-early theory published around the same time is closely coupled to the exon theory of genes [5][6][7] [5–7]. The main hypothesis is that the split structure of genes is ancient and occurs in all domains of life, and that exons are ancestral genetic elements encoding proteins with small domains. Thus, multidomain proteins evolved by recombination of neighboring exons that were connected by noncoding sequences [8][9][8,9]. Subsequently, these noncoding sequences became introns. However, further investigations showed inconsistent correlation between exons and protein domain units [10].
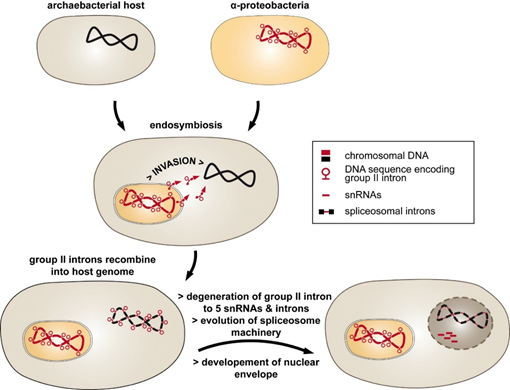
An alternative, the introns-late theory, was proposed by different authors [11][12][11,12]. According to this hypothesis, spliceosomal introns are unique to eukaryotes, introduced into the host genome by an endosymbiont (Figure 1). The endosymbiosis of α-proteobacteria entailed massive genomic transfer accompanied by invasion of the host genome with group II introns. This type of intron has the ability to self-splice and invade DNA, and it is found in abundance in eubacterial genomes [13][14][13,14]. Strong selective pressure on the archaeal host may have initiated the fragmentation of group II intron RNA into five small nuclear RNAs (snRNAs) and the mRNA intron, which now function in splicing of spliceosomal introns [15]. Consistent with this idea of intron evolution is the distribution of group II introns [13]. They are frequently found in bacterial genomes and were most likely introduced by horizontal transfer to eukaryotic nuclei [16].
Figure 1. The introns-late theory explains the evolution of spliceosomal introns. The endosymbiosis of a group II intron-rich α-proteobacteria into the archaeal intronless host was followed by the invasion of the host genome by mobile group II introns. The resulting discontinuous genes provoked a strong selective pressure toward evolving intron removal. This included the degradation of group II intron RNA into mRNA introns and small nucleolar RNAs (snRNAs). Separation of the inefficient splicing reaction from translation was achieved by developing a nuclear envelope.
So far, group II introns have not been found in nuclear genomes of eukaryotes, which indicates that they were efficiently transformed into spliceosomal introns during evolution. The introns-late theory was further expanded by proposing that the invasion of group II introns was one of the major driving forces for nucleus–cytoplasm compartmentalization [17]. Accordingly, the “turbulent phase” of gene transfer, including the spread of group II introns throughout the host genome, was followed by the development of low-efficiency splicing reactions. These circumstances led to the accumulation of unspliced precursor RNAs and translation of non-sense proteins. Thus, the formation of a nuclear envelope separated RNA splicing and protein translation within the cell.
Overall, the evolutionary development of spliceosomal introns delineated in the introns-late theory seems to be most likely. However, more parallels exist between group II introns and their putative counterparts in the nuclei of eukaryotes, as we discuss in the following sections.
3. Group II Introns and Spliceosomal Introns Share Identical Splicing Mechanisms
One of the most striking parallels of spliceosomal and group II introns is their identical splicing mechanism [18] [11,18]. Current textbooks describe in detail the two transesterification reactions that occur during RNA splicing of group II introns. One important intermediate arising during this process is a lariat-formed RNA molecule that resembles the intron stem-loop lariat catalyzed by the spliceosome during nuclear pre-mRNA splicing. Thus, the formation of an intron lariat is a conserved feature of both the spliceosomal and group II intron splicing reaction.
In vitro, the splicing mechanism seems to be a simple chemical reaction, but in vivo, the process requires an intricate network of RNA–RNA interactions which are stabilized by protein factors [19][20][21][14,19–21]. The initial step during splicing is the recognition of exon–intron boundaries and the branch point nucleotide. Next, the 5′ss, 3′ss, and branch point are juxtaposed to create the catalytic core, where they are surrounded by further catalytic RNA structures and become reactive. These steps are performed by conserved RNA sequences and structures.
4. Sequence Homologies and Mechanistic Similarities
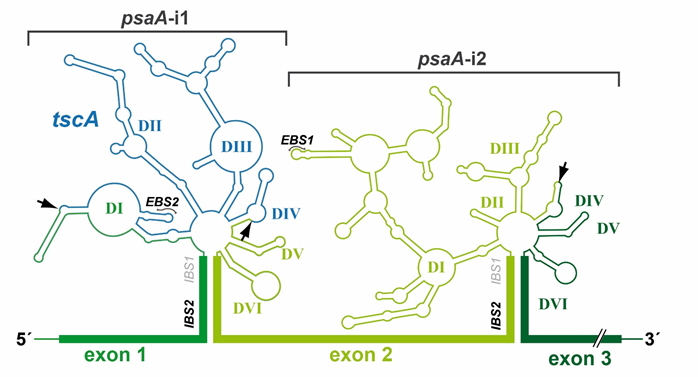
All group II introns display a characteristic and highly conserved secondary structure, (Figure 2). A central core containing the splice sites is surrounded by six helical domains (DI-DVI). Sequence similarities are rare and only occur in catalytic structures [22]. RNA–RNA interactions between intron sequences assemble a conserved tertiary structure that forms the catalytic core [23][24][23,24]. Deletion analysis showed that minimal catalytic activity requires at least DI and DV [25]. DII and DIII enhance activity but are not essential for accurate splicing [26][27][26,27]. Finally, DIV harbors an optional open reading frame (ORF) encoding a maturase [28]. These proteins comprise three distinct domains: an RNA-binding and splicing domain (X), a reverse transcriptase (RT) domain, and a DNA-binding/endonuclease (D/En) domain. The latter mediates the mobility of group II introns.
Figure 2. Secondary structure of psaA-i1 and psaA-i2 intron RNAs from C. reinhardtii. The three exons of the psaA gene and tscA locus of C. reinhardtii are transcribed independently. Two group II introns are formed at the exon boundaries of psaA by RNA base pairing. The mature psaA mRNA is generated by two trans-splicing reactions. Both group II introns have a characteristic group II intron structure, comprising six helical domains (DI-DVI), which surround a central core. psaA-i1 is a tripartite intron, whereas psaA-i2 is a dipartite intron. Fragmentation sites are indicated by arrows. Abbreviations: EBS/IBS, exon-/intron-binding sites.
In contrast to group II introns, splicing of nuclear mRNA introns requires the intron sequence itself and trans-acting RNAs. The pre-mRNA intron harbors the 3′/5′ss and the branch point nucleotide. However, besides the intron sequence, splicing of nuclear mRNA introns requires five trans-acting RNAs, the snRNAs. The snRNAs assemble with protein factors and form ribonucleoprotein (RNP) complexes [29] [29]. For each splicing reaction, these RNPs associate de novo on one intron together with a further set of more than 100 proteins and the intron RNA [19]. This highly dynamic RNP splicing machinery is known as the spliceosome.
According to the introns-late theory, snRNAs originated from group II intron sequences. Indeed, similar to the helical domains of group II introns, they mediate exon/intron recognition and catalytic core formation [30].
The detection of introns is also crucial for splice site recognition, mainly because they are located directly at the boundaries of exon sequences. Consistent with the introns-late theory, group II introns and spliceosomal introns share sequence similarities at their intron ends. Group II introns start with 5′-GUGYG and end with AY-3′ (Y = C, U), while nuclear U2-type mRNA intron boundaries comprise 5′-GU-AG-3′ [14]. The structural similarities between group II introns and spliceosomal introns and their similar chemistry of splicing were excellently discussed recently [31][32][31,32].
Besides recognizing regions defining exon and intron boundaries, efficient splicing of group II and nuclear mRNA introns requires a bulged nucleotide, which is involved in the first nucleophilic attack. With group II introns, this nucleotide is generally an adenosine located in domain DVI (Figure 2). In spliceosomal introns, the branch point emerges close to the 3′ss after binding of U2 snRNP to the mRNA intron sequence [33][34][33,34]. This recognition is maintained by a consensus sequence and binding results in a single bulged nucleotide. The common fixation of a bulged nucleotide conformation is a further indication for the evolutionary relationship of both intron types [35].
The catalytic core is the most striking parallel between group II and spliceosomal introns [36][37][23,36,37]. It requires the 3′/5′ss and branch point nucleotide to be juxtaposed in a reactive manner and is characterized by further catalytic RNA structures and sequences [38][36,38]. In group II introns, the catalytic core is defined by the interaction of two distinct structures, the junction between DII and DIII (J2/3) and DV [39][14,39] (Figure 2). DV is the most phylogenetically conserved structure in group II introns and, therefore, is often described as the heart of a group II intron [40][38,40]. It comprises two short helices separated by a bulged region and contains the catalytic triad (5′-RGC, R = A/G) crucial for binding of J2/3. This interaction results in a triple helix structure [41]. The DV bulge (Figure 2) is mainly responsible for metal ion binding, which is required for efficient splicing [41] [35,41].
The equivalents in the spliceosome are found in the U6/U6atac snRNAs [42][43][37,42,43]. The evolutionary conserved elements are the ACAGAGA box corresponding to J2/3 and an AGC triad in a bulged internal stem loop that corresponds to domain DV. The importance of these structures was supported by mutation analyses of the corresponding sequences, resulting in complete or partial loss of splicing efficiency [44][45][46][44–46]. These remarkable similarities in RNA structure and functionality provide further hints that snRNAs originated from group II introns.
5. Trans-Spliced and Degenerated Group II Introns: A Step Toward Spliceosomal Introns?
The origin of trans-acting snRNAs from fragmentation of ancestral group II introns is a reasonable theory to explain the evolution of spliceosomal introns. However, neither intermediate stages of this fragmentation process nor sequences encoding group II introns have been identified so far in more than 1000 sequenced nuclear genomes [47][48][14,47,48]. Recently, group II introns artificially inserted into nuclear genes of Saccharomyces cerevisiae were shown to be expressed and spliced [49][50][49,50]. However, the group II introns were not spliced until they were transported into the cytoplasm, in contrast to spliceosomal introns that are spliced exclusively in the nucleus. Additionally, translation of group II intron-spliced mRNA was inhibited due to translational repression.
The search for intermediate states in the transition of group II introns to nuclear mRNA introns therefore focused on cell compartments (chloroplasts, mitochondria) and bacteria that still contain group II intron-encoding sequences. Indeed, discontinuous group II introns were identified in organellar genomes and were used to illustrate what could have occurred during evolution [51][52][15,47,51,52]. As a consequence of genome rearrangements, exons of genes encoding such fragmented group II intron structures are dispersed across the eukaryotic genome [53]. The independently transcribed precursor RNAs associate through base pairing and tertiary interactions to generate the catalytic conserved group II intron structure. Therefore, mature mRNA is generated by trans-splicing, in contrast to the process of cis-splicing that involves a single continuous RNA precursor.
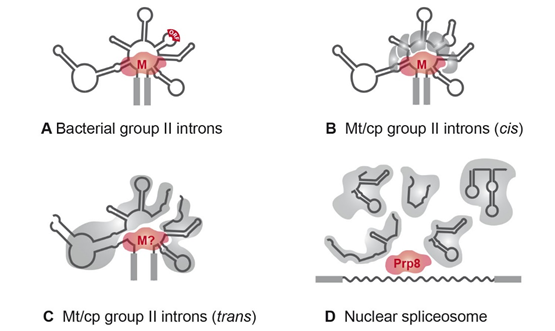
The first organellar trans-spliced group II introns were identified in the late 1980s [54][55][56][54–56]. The majority of trans-split group II introns are dipartite and disrupted in domain DIV [57][52,57]. This domain is insufficient for proper splicing, but it harbors an optional ORF encoding a maturase (Figure 3A). The fragmentation is consistent with the observation that nearly all organellar group II introns have lost the maturase-encoding gene [58][20,58]. If trans-spliced group II introns still harbor an ORF, a fragmentation site is located either upstream or downstream of the ORF [59]. Hence, fragmentation of the group II intron structure in DIV occurs quite often, while other domains are less affected (Figure 3C).
Figure 3. RNPs of group II and spliceosomal introns. (A) The majority of bacterial group II introns form an RNP with the intron-encoded maturase during the splicing reactions [14]. (B) In organelles, group II introns are degenerated and RNPs comprise at least five splicing factors [20]. The maturase is either encoded by an organellar group II intron (matR, matK) sequence or is nucleus-encoded (nMat1–4; [28]). (C) Fragmented group II introns of C. reinhardtii depend on complex RNPs comprising up to ten splicing factors and the precursor RNAs. A maturase homolog has not been identified yet. (D) The five trans-acting snRNAs of the nuclear spliceosome probably originated from fragmentation of group II intron sequences. snRNAs associate with a large number of protein factors to form a complex with snRNPs and function in splicing of nuclear mRNA introns (reviewed in [4]). Homologies to group II intron maturases were shown for the splicing factor Prp8 [60]. Abbreviations: M, maturase; Mt, mitochondrial; cp, chloroplast.
Two dipartite group II introns were shown to be disrupted in DIII and are located in the rps12 gene of Marchantia and Nicotiana [61][54,61]. Additionally, the rbcL-intron 2 (rbcL-i2) of the green algae Floydiella terrestris and Stigeoclonium helveticum is disrupted in DII [62][63][62,63]. RNA elements of the helical DII and DIII structures are also not sufficient for splicing [64]. Furthermore, some dipartite group II introns (rbcL-i1, psaC-i1, and petD-i1) in plastids of a few green algae show fragmentation in DI [65][62,63,65].
Additional fragmentation of group II introns leading to a tripartite structure is rare, with two representative examples: the chloroplast intron psaA-i1 in the green alga Chlamydomonas reinhardtii and the mitochondrial intron nad5-i4 in the flowering plant Oenothera berteriana [56,66]. Both introns exhibit one disruption in DIV and one in DI, which occurs close to the ε-binding site (Figure 2). Thus, the EBS1/δ-containing structure of domain DI, DII, and DIII must be reconstituted by a third RNA fragment. In both organisms, loci encoding such trans-acting RNA structures have been identified and are located in distant genomic regions. These are the tscA (trans-splicing of chloroplast psaA mRNA) locus in C. reinhardtii and the tix (trans-splicing intron fragment) locus in O. berteriana [66][67][66,67]. Surprisingly, structural predictions indicate that the tscA RNA provides only a degenerated DI lacking EBS1 and δ-interaction sites. The absence of these structures important for the exon recognition process leads to the question of whether a fourth RNA is involved in psaA-i1 splicing [67].
The ability of natural continuous group II introns to trans-splice was previously investigated using a transposon-based genetic screen [68][69] [51,68,69], where dipartite and tripartite versions of the Ll.LtrB group II intron from Lactococcus lactis were generated to analyze their capacity to splice. The Ll.LtrB intron proved to be remarkably tolerant to fragmentation and able to splice when such fragmentation corresponded to naturally occurring sites in DI, DIII, and DVI. In contrast, fragmentation was not tolerated within functionally important elements, such as EBS1/EBS2, DVI, and structures involved in central core formation, including the splice sites.
In a recent approach, an alternatively spliced group II intron was even identified in the bacterial pathogen Clostridium tetani [70]. The C.te.I1 intron is located in the surface layer gene (SLP) and is able to undergo four alternative splicing reactions in vivo, thus producing different isoforms of SLP necessary for virulence and resistance to the host immune response. Alternative splicing is common in spliceosomal introns. Approximately 95% of human genes are thought to be alternatively spliced [71]. This process leads to a dramatic increase in information encoded by the genome [72][73][72,73]. The fact that group II introns are also used in this advantageous manner emphasizes their functional similarities with spliceosomal introns.
Another process indicating a transition toward nuclear mRNA introns includes deletions within the intron sequence. All mitochondrial and plastid group II introns of higher plants have lost the ability to self-splice, in contrast to bacterial group II introns [74]. This is primarily due to a degenerated or completely lost maturase gene in DIV [75][76][75,76]. However, several other alterations have been observed. For example, some mitochondrial group II introns in land plants display an abnormal DVI structure and lack a bulged adenosine, as was shown for nad1-i1, nad1-i2, nad4-i2, rpl2, and rps3 [77][47,77]. The homologous intron of nad4-i2 in the green alga Chara vulgaris exhibits a common DVI structure [78 [78]. This emphasizes that alterations in higher plants occurred by rearrangements of a classical group II intron structure.
The occurrence of split group II introns in organellar eukaryotic genomes demonstrates that degeneration and fragmentation is possible and might represent an intermediate stage in the evolution of spliceosomal introns. Furthermore, the identification of trans-acting RNAs during organellar group II intron splicing illustrates how spliceosomal snRNAs may have originated.