Copper is an essential transition metal which is also toxic to cell.. Organisms have developed sophisticated pathways to import, traffic, store and deliver copper to cuproproteins. They also export its excess outsite of the cell to protect themselves from oxidative stress. The pathways contains specific importers, chaperons, storage proteins and exporters. Expression of the corresponding structural genes is conrolled by copper availability via sensors and response regulation transcription factors described below.
- copper
- bacteria
- cuprenzymes
1. Introduction
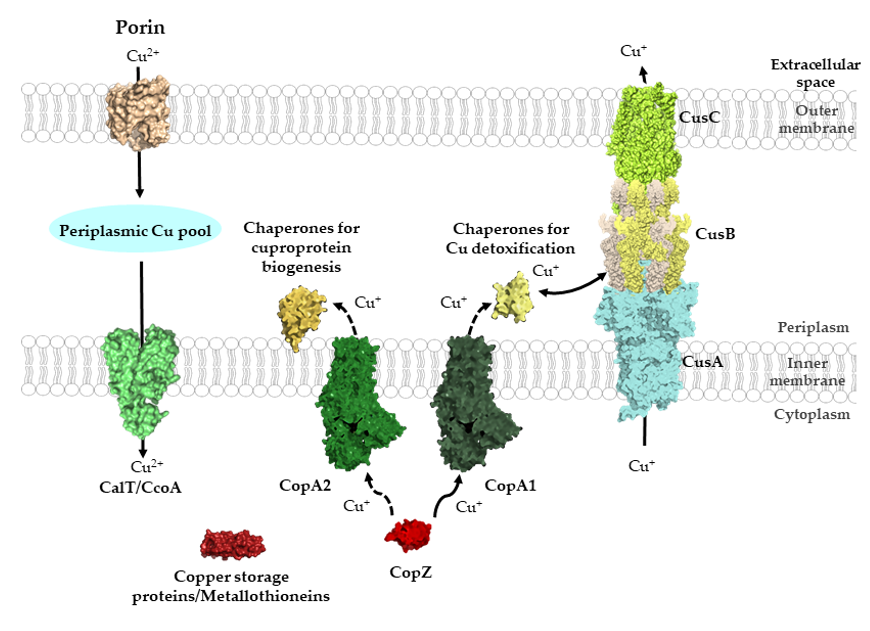
Approximately 30% of all proteins in bacteria depend on metals for their function. Understanding how these potentially toxic metals are imported into bacterial cells, and how they are ultimately delivered to their target proteins without inducing toxic effects is a crucial issue in metalloprotein biogenesis (Figure 1) [1,2][1][2]. In contrast to most other nutrients, the concentration of many metals in natural environments usually exceeds cellular needs and hence multiple mechanisms that prevent metal-induced damage are encountered in bacteria. Copper (Cu) is one such metal that is essential in eukaryotes and prokaryotes but is highly toxic when present in excess. Indeed, Cu alloys are used for controlling bacterial surface contaminations [3]. Furthermore, human macrophages pump copper into their phagosomes after engulfing pathogenic bacteria to induce oxidative stress and bacterial cell death [4]. Consequently, bacteria have to deal with high copper concentrations in order to evade the host immune system [4–7][4][5][6][7]. The importance of Cu detoxifying systems for bacterial virulence is exemplified by the observation that the inactivation of Cu-exporting P1B-type ATPases in Mycobacterium tuberculosis impairs their ability to proliferate in host macrophages [8,9][8][9]. The link between Cu homeostasis and bacterial virulence is summarized in several recent reviews and not covered in depth here [5,10–15][5][10][11][12][13][14][15].
Cu toxicity is intrinsically linked to its redox properties that favor the generation of reactive oxygen species via a Fenton-like reaction:
|
Cu+ + H2O2 → Cu2+ + OH− + .HO |
|
In addition, Cu is located on top of the Irving-Williams series and Cu binding to proteins is usually a thermodynamically favored process [16]. Although this aids Cu insertion into cuproenzymes, excess Cu could lead to significant mis-metalation of proteins naturally containing other metals, and iron in particular, resulting in their inactivation [17–19][17][18][19]. Potent targets of Cu toxicity in the periplasmic space of bacteria are the biogenesis pathways for cytochrome c [19] and bacteriochlorophyll synthesis [20], aside from interfering with the thiols of periplasmic proteins such as the thiol:disulfide oxidoreductases [21].
Figure 1. General view of Cu transport across bacterial membranes. Cu can cross the outer membrane of bacteria via porins and major-facilitator superfamily members, such as CcoA, can import periplasmic Cu into the cytosol. Additional Cu importers likely exist but have not been characterized in detail. Cytosolic Cu is bound by Cu storage proteins, metallothioneines and Cu chaperones, such as CopZ. Cu-chaperones also deliver Cu to P1B-type ATPases for export into the periplasm. Kinetic differences distinguish CopA1-like ATPases, which are involved in Cu detoxification, and CopA2-like ATPases, which export Cu for cuproenzyme biogenesis. CopA1-like ATPases are the primary interaction partner of CopZ, while the interaction with CopA2-like ATPases is particularly important at low Cu concentrations (dashed arrow). In the periplasm, different types of chaperones transfer Cu either for cuproenzyme biogenesis or for Cu export systems, as with the CusABC system. The structures shown were retrieved from the protein database (PDB) with the following IDs: 2ZFG (OmpF for Porin), 3WDO (YajR for CalT/CcoA), 5NQO (Csp3 for copper storage proteins), 1K0V (CopZ), 3j09 (CopA1, CopA2) 4WBR (ScoI/SenC, chaperones for cuproprotein biogenesis), 2VB2 (CusF, chaperones for Cu detoxification), 3KSS (CusA), 3H94 (CusB), 4K7R (CusC), and are depicted using Pymol
The high redox potential of the Cu(II)/Cu(I) pair (+160 mV) favors reactions with oxygen and oxygen containing molecules and it is generally assumed that Cu-dependent proteins have evolved concomitantly with the appearance of molecular oxygen, which started about 3 × 109 years ago [22]. Studies on Cu transport have mainly focused on Cu export for Cu detoxification, in line with its harmful effects, while the mechanisms of Cu import across the outer membrane of Gram-negative bacteria and the bacterial cytoplasmic membrane have been analyzed in only a few cases (Figure 1) [23,24][23][24].
Cu-binding proteins (cuproproteins) in the bacterial cytosol act mainly as Cu-chaperones, Cu storage proteins and Cu-responsive transcriptional regulators [25]. In contrast, cuproenzymes, which contain Cu as part of their catalytic center and bind Cu permanently, are primarily involved in aerobic and anaerobic electron transfer reactions, monooxygenation reactions or superoxide dismutation (Table 1). Intriguingly, except plastocyanin, located in the thylakoid lumen [26] and required for electron transport processes, most cuproenzymes identified so far are localized to the bacterial membrane, the periplasmic space or the cell surface. The almost complete absence of cytosolic cuproenzymes in bacteria might be one strategy to prevent Cu toxicity simply by compartmentalization. This is also exemplified by the fact that almost all studied cuproenzymes appear to be metalated outside of the cytosol [25,27,28][25][27][28]. Even for the periplasmic multi copper oxidase CueO, which folds partially inside of the cytosol and is secreted by the Tat protein transport pathway [29], Cu insertion likely occurs in the periplasm [30]. However, bacterial cells contain a cytosolic Cu pool of often poorly defined nature. For example, detailed studies on the Cu delivery pathway for the cbb3-type cytochrome c oxidase (cbb3-Cox), have demonstrated that assembly and activity of cbb3-Cox is strictly dependent on the P1B-type Cu-exporting ATPase CcoI [31,32][31][32]. Thus, even though Cu is inserted into the catalytic heme b-CuB center of cbb3-Cox from the periplasmic side of the membrane [33], Cu delivery still depends on an obligatory cytosolic Cu pool. One possible explanation for this puzzling observation is that Cu is preferentially inserted into cuproenzymes as chaperone-bound Cu(I) and routing it through the cytoplasm, ensures control of its reduced state as Cu(I).
Table 1. Examples of bacterial cuproenzymes. Listed are cuproenzymes that stably bind copper in their catalytic center. Cuproproteins, which only transiently bind copper, such as Cu-chaperones, Cu-binding proteins or Cu-responsive transcriptional regulators, are described in the main text.
|
Protein |
Localization |
Function |
Reference |
|
Plastocyanin |
Thylakoid lumen |
Photosynthetic electron transfer |
[26] |
|
Cytochrome c Oxidase |
Membrane |
O2-reduction |
[34] |
|
Particulate Methane monooxygenase |
Membrane- associated |
Methane hydroxylation |
[35] |
|
Multi-Copper oxidases |
Periplasm |
Cu detoxification |
[36] |
|
Nitrite reductase |
Periplasm |
Nitrite reduction |
[37] |
|
Azurin |
Periplasm |
Respiratory electron transfer |
[38] |
|
Cu-Zn Superoxide dismutase |
Periplasm |
Superoxide detoxification |
[39] |
|
Nitrous oxide reductase |
Periplasm |
Denitrification |
[40] |
|
Amine oxidases |
Periplasm |
Amine oxidation |
[41] |
|
MccA-type Sulfite reductase |
Periplasm |
Sulfite reduction |
[42] |
|
Tyrosinase |
Extracellular |
Monooxygenase |
[43] |
Cu insertion into cuproenzymes is generally facilitated by dedicated auxiliary Cu-chaperones, such as NosL for nitrous oxide reductase [44,45][44][45] or the ScoI- and PCuAC-like chaperones for Cox [46,47][46][47]. Intriguingly, in some cases, these Cu-chaperones can be bypassed by increasing the Cu-concentration in the medium, suggesting that they are particularly important at low Cu availability. In bacteria, Cu-chaperones can also act on different targets. The ScoI- and PCuAC-like chaperones were initially linked to the assembly of the periplasm-exposed binuclear CuA site in subunit II of aa3-Cox [48–51][48][49][50][51]. However, it is now evident that both proteins are also involved in the formation of the deeply membrane-buried CuB-site of subunit I of cbb3-Cox [46,47,52,53][46][47][52][53].
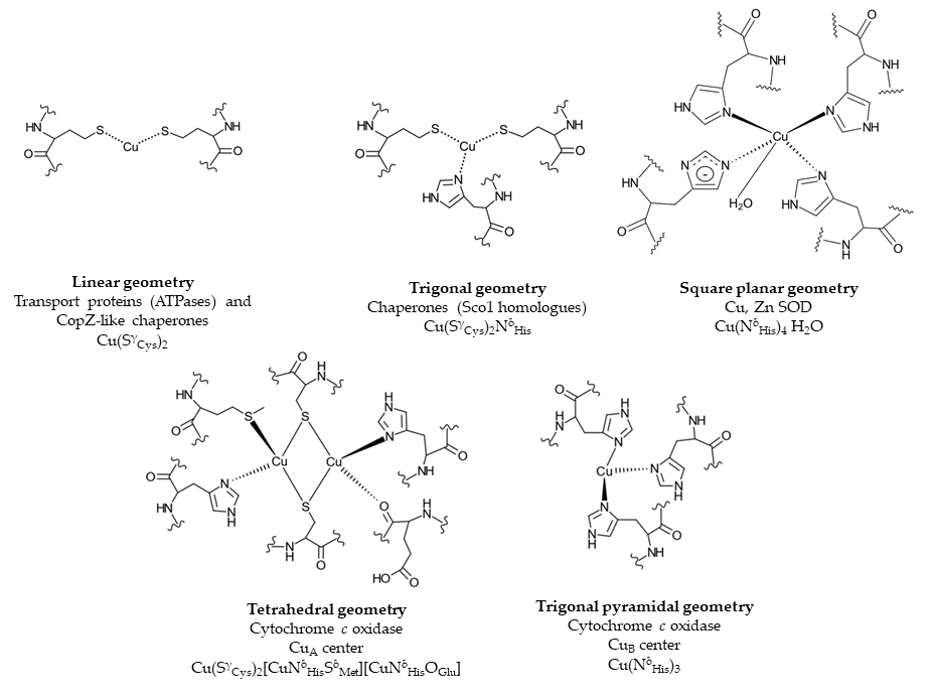
Cu binding sites within proteins are usually composed of cysteine, methionine, histidine and occasionally tryptophane residues [54]. The composition and geometry of the Cu binding motif is important for its reduction potential. While redox cycling is required for the catalytic activity of cuproenzymes, cuproproteins involved in Cu trafficking need to avoid Cu(I)/Cu(II) redox cycling and therefore their Cu binding motifs require a more negative reduction potential that stabilizes the Cu(I) state [54] (Figure 2). Copper trafficking proteins coordinate Cu(I) via a CxC or CxxC motif with a linear geometry as in Cu(I)-transporting P1B-type ATPases or in CopZ-like Cu chaperones [54], or with a trigonal planar geometry as in Sco1-like chaperones. Here, two cysteine residues and a distal histidine residue provide the binding site for Cu(I). In cuproenzymes, such as Cu,Zn superoxide dismutase (SOD) or cytochrome oxidase, Cu is coordinated by histidine, cysteine and methionine residues (Figure 2). Cu(II) is preferentially liganded via histidine nitrogen donors in a square planar arrangement [55–57][55][56][57]. Methionine as ligand is less pH sensitive and more hydrophobic and provides often an additional weaker ligand for Cu [58].
Figure 2. Examples of Cu binding sites in Cu trafficking proteins and cuproenzymes. Cu-transporting P1B-type ATPases and Cu chaperones, such as CopZ, contain a linear CxxC Cu binding motif, while in Cu chaperones, e.g., Sco1, a distal histidine residue provides an additional ligand, resulting in a trigonal planar geometry. In the cuproenzyme Cu, Zn superoxide dismutase (SOD), four histidine residues ligate Cu in a square planar geometry. The CuA center of cytochrome oxidase displays a tetrahedral geometry with two cysteines, two histidines, one glutamate and one methionine residue. In the CuB center, Cu is ligated by three histidine residues. Chemical structures were generated using ChemDoodle. The figure was adapted and modified from [54].
2. Copper Import across the Outer and Inner Membranes in Bacteria
Since Cu is an essential micronutrient, its passage across biological membranes is crucial for all organisms. In eukaryotes, Cu import is mainly mediated by members of the Ctr (copper transport) family of transporters [59]. Eukaryotic cells contain between one and six Ctr family members (also known as SLC31 family), which display some subcellular or organ specificity [59]. Especially Ctr1 is well characterized in terms of its physiology, structure and function [60–63][60][61][62][63]. Cytosolic Cu is further distributed into eukaryotic organelles by P1B-type ATPases (also known as CPx-ATPases) [64]. In contrast to the Ctr family members, which so far have not been identified in bacterial genomes [62], P1B-type ATPases are universally conserved and widely distributed in different bacterial species [1]. As in eukaryotes, bacterial P1B-type ATPases export Cu out of the cytosol and are apparently not involved in Cu import. Although some P1B-type ATPases, such as CtaA of the cyanobacterium Synechocystis PCC 6803 and CopB of Enterococcus hirae, were initially suggested to import Cu into the cytosol, further analyses indicated that both proteins are involved in Cu export out of the cytosol [65–68][65][66][67][68]. In cyanobacteria, a second P1B-type ATPase, PacS, distributes Cu further into the thylakoid lumen for plastocyanin and caa3-Cox assembly [69–71][69][70][71].
Bacterial Cu importing systems in general are more diverse than in eukaryotes and some of the well-studied mechanisms of Cu uptake are limited to one group of bacteria, or even to one species. This section will mainly focus on Cu import mechanisms in Gram-negative bacteria, where most of the well-studied Cu-uptake pathways have been described.
2.1. Cu-Uptake across the Outer Membrane of Gram-Negative Bacteria and Mycobacteria
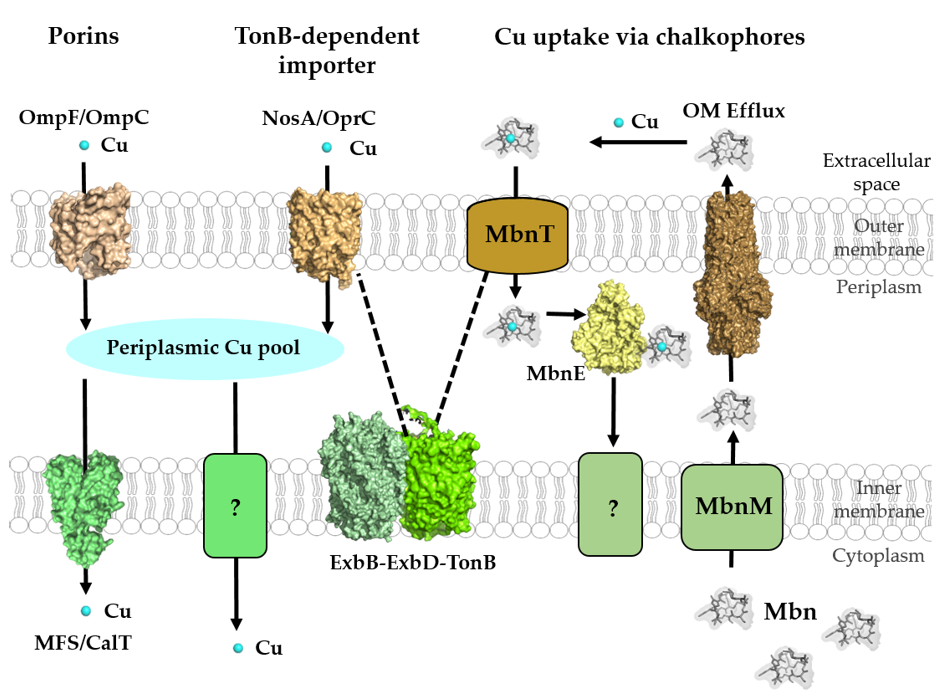
The passage of ions and small molecules across the outer membranes generally occurs either via various porins or through energy-dependent mechanisms. Transcriptional profiling and comparative differential cuproproteomics using Pseudomonas aeruginosa [72], Acidithiobacillus ferrooxidans [73] and Rhodobacter capsulatus [74] revealed that the expression or steady-state levels of several outer membrane proteins changed when cells were exposed to Cu stress. For most of these proteins, a role in Cu uptake still remains to be defined, but some studies have identified specific outer membrane proteins with more defined role(s) in Cu uptake (Figure 3).
- coli mutants lacking the outer membrane porin OmpF were shown to be Cu resistant, suggesting a role of this porin in Cu(II) uptake [75]. However, various isogenic ompF and ompC mutants of other E. coli strains did not show significant changes in Cu resistance, suggesting the presence of additional Cu import systems [76]. The involvement of porins in Cu uptake has also been shown in mycobacteria, which contain a complex cell envelope that consists of the cytoplasmic membrane, the peptidoglycan–arabinogalactan complex and an outer membrane that is covalently attached to arabinogalactan [77,78]. In Mycobacterium smegmatis, mutants lacking the porins MspA or MspC show severe growth defects on Cu-limited growth media, and increased Cu tolerance when grown at high Cu concentrations [79]. Heterologous production of M. smegmatis MspA suppresses growth of M. tuberculosis at high Cu concentration, further indicating that MspA is involved in Cu uptake across their outer membrane [79–81]. The X-ray diffraction analysis of M. smegmatis MspA showed an interconnected octamer with eightfold symmetry that resembles a goblet with a single central channel [80]. A recent study, searching for new strategies to boost the antimicrobial activity of Cu, identified 8-hydroxyquinoline (8HQ) as a potent Cu-dependent bactericidal of M. tuberculosis. However, the antimicrobial activity of 8HQ–Cu was also observed in a ΔmspA mutant strain of M. smegmatis, suggesting that MspA is not essential for the uptake of the 8HQ–Cu complex [82].
E. coli mutants lacking the outer membrane porin OmpF were shown to be Cu resistant, suggesting a role of this porin in Cu(II) uptake [75]. However, various isogenic ompF and ompC mutants of other E. coli strains did not show significant changes in Cu resistance, suggesting the presence of additional Cu import systems [76]. The involvement of porins in Cu uptake has also been shown in mycobacteria, which contain a complex cell envelope that consists of the cytoplasmic membrane, the peptidoglycan–arabinogalactan complex and an outer membrane that is covalently attached to arabinogalactan [77][78]. In Mycobacterium smegmatis, mutants lacking the porins MspA or MspC show severe growth defects on Cu-limited growth media, and increased Cu tolerance when grown at high Cu concentrations [79]. Heterologous production of M. smegmatis MspA suppresses growth of M. tuberculosis at high Cu concentration, further indicating that MspA is involved in Cu uptake across their outer membrane [79][80][81]. The X-ray diffraction analysis of M. smegmatis MspA showed an interconnected octamer with eightfold symmetry that resembles a goblet with a single central channel [80]. A recent study, searching for new strategies to boost the antimicrobial activity of Cu, identified 8-hydroxyquinoline (8HQ) as a potent Cu-dependent bactericidal of M. tuberculosis. However, the antimicrobial activity of 8HQ–Cu was also observed in a ΔmspA mutant strain of M. smegmatis, suggesting that MspA is not essential for the uptake of the 8HQ–Cu complex [82].
NosA is an outer membrane protein that is involved in the biogenesis of the periplasmic, Cu-containing enzyme nitrous oxide reductase (NosZ). NosZ contains two Cu centers (CuA and CuZ), which are required for N2O reduction [83,84][83][84]. NosA was first identified in Pseudomonas stutzeri in a series of mutants producing inactive NosZ variants that lacked Cu in the catalytic center [85]. NosA shows some homology to known E. coli TonB-dependent outer membrane transport proteins, including BtuB, TutA, FepA and FhuA [86]. TonB-dependent transport systems are unique in bacteria and use the electrochemical potential of the cytoplasmic membrane for transporting nutrients across the outer membrane [87–90][87][88][89][90]. NosA is likely required for NosZ assembly at low Cu concentrations and might also function as a general Cu import system, rather than dedicated specifically to NosZ assembly [91,92][91][92]. Nonetheless, NosA-dependent Cu uptake is still considered as one of the main routes for Cu delivery to NosZ in NosA-containing organisms, but alternative pathways for NosZ metalation are suggested to exist in organisms lacking NosA [18].
Figure 3. Cu-transport across the outer and inner membranes of bacteria. Bacteria use three established systems for Cu transport across the outer membranes: porins, TonB-dependent transport systems, such as NosA/OprC, and uptake via chalkophores, such as Methanobactin (Mbn), which represent Cu-specific metallophores. Periplasmic Cu is transported into the cytoplasm via the CcoA-like transporter (CalT), which belongs to the major facilitator superfamily. Additional Cu import systems (e.g., YcnJ-like proteins [93]) likely exist in bacteria, but have not been characterized [23]. Mbn is synthesized in the cytoplasm and transported via MbnM into the periplasm and by a so far unknown transporter into the extracellular space. Cu-loaded Mbn is imported back into the periplasm via the TonB-dependent transporter MbnT and bound by the periplasmic protein MbnE. How Cu-Mbn is transported back into the cytosol across the cytoplasmic membrane and how Cu is released from Mbn in the cytosol are unknown. Dashed lines indicate the interaction of the TonB-dependent transporter NosA/OprC and MbnT with the inner membrane ExbB–ExbD–TonB complex. Structures were retrieved from the protein database (PDB) with the following IDs: 2ZFG (OmpF for Porin), 3WDO (YajR for CalT/CcoA), 3j09 (CtaA), 4ZA1 (NosA), 5ZFP (ExbB/ExbD hexameric complex), 5ICQ (MbnE), 1EK9 (OM efflux), 2XJH (Mbn), and depicted using Pymol.
OprC is a TonB-dependent transporter in the outer membrane of Pseudomonas aeruginosa. OprC is homologous to NosA of P. stutzeri (65% amino acid sequence identity) and binds Cu(II) with micromolar affinities [94]. The expression of OprC was shown to be repressed by high exogenous Cu(II) concentrations and enhanced under anaerobic conditions in the presence of nitrate [94,95][94][95]. Although, OprC expression is repressed under Cu stress, it is induced by low Cu(II) concentrations and is directly regulated via the Cu-responsive transcriptional regulator CueR in P. aeruginosa [96]. Moreover, OprC is an important determinant for bacterial competition and virulence [96]. Very recently, the crystal structures of OprC wild-type and mutant proteins were resolved in the presence and absence of silver and Cu [97]. The structures. as well as the inductively coupled plasma mass spectrometry (ICP-MS) and electron paramagnetic resonance (EPR) data, suggested that Cu(I) binds to a CxxxM-HxM motif. It was furthermore suggested that OprC also binds Cu(II) and is able to reduce it to Cu(I) via thiol groups, although this awaits further validation [97].
Unlike NosA and OprC, the small outer membrane protein ComC (also known as YcfR or BhsA) acts by lowering the permeability barrier of the outer membrane to Cu. Most Gram-negative bacteria encode ComC-like proteins with 50% to 90% sequence homology. In the absence of ComC, E. coli shows reduced Cu import into the cytoplasm [98]. The transcription of comC is induced via the TetR-like transcriptional regulator ComR in response to Cu availability [67,98][67][98]. Initially, ComC was described as a general stress response protein [99–102][99][100][101][102] and it remains unclear how ComC is simultaneously involved in different cellular stress response pathways, including Cu stress.
2.2. Cu Transit through the Periplasmic Space in Gram-Negative Bacteria
Once Cu has crossed the outer membranes into the periplasm, it is likely scavenged by periplasmic Cu chaperones, Cu-storage proteins and chemical chelators, such as glutathione [18,103,104][18][103][104]. Although several periplasmic Cu chaperones have been described in multiple bacteria, they are mainly associated with Cu-detoxification and cuproenzyme biogenesis pathways [47,52,105][47][52][105]. Hence, it is currently not clear, whether they also participate in Cu uptake, and therefore these proteins are described later. Dedicated Cu-storage proteins (Csps) were first described in the Gram-negative methanotrophic bacteria, but are widely distributed in bacteria [103]. Csp1-type proteins contain a Tat signal sequence and are likely exported into the periplasm for Cu binding [103,106,107][103][106][107]. As cytosolic Csp3-type proteins are much more abundant than the secreted Csp1-type proteins [103], they are further discussed in Section 4.4.
2.3. Cu Uptake across the Inner Membrane
The CcoA-Like Cu-Transporter (CalT) Family
Major-facilitator-superfamily (MFS)-type transporters belong to a large and ubiquitous superfamily of transporters. MFS proteins selectively transport a wide range of substrates by using the proton gradient as driving force [108]. CcoA belongs to the newly discovered CalT (CcoA-like transporter) family of MFS-type transporters (Figure 1), and is the prototype of a bacterial inner membrane Cu importer [23,33,109,110][23][33][109][110]. CcoA is the first Cu uptake transporter identified in bacteria and the first MFS-type transporter known to transport Cu. It was first discovered in R. capsulatus by genetic screening for cbb3-Cox defective mutants that could be rescued by exogenous Cu supplementation. Furthermore, heterologous expression of ccoA restores the respiratory defect and Cu import in a Schizosaccharomyces pombe double mutant that lacked the Cu-importer Ctr4 and Ctr5 [111]. Overall, these studies demonstrated that CcoA is a Cu importer required for the biogenesis of R. capsulatus cbb3-Cox. Interestingly, bypass suppressors of ccoA deletion mutants that suppress the cbb3-Cox defect were frequently observed in R. capsulatus [23,109][23][109]. These bypass suppressors were Cu sensitive and had higher cellular Cu content compared to wild-type and ccoA mutant strains. Whole genome sequencing revealed that these suppressor mutants contained single base pair insertion/deletion in copA [109], encoding the well-known P1B-type Cu exporter, CopA, involved in Cu detoxification [25,112][25][112]. These observations indicated the presence of a functional interplay between the Cu importer, CcoA, and the Cu exporter, CopA, in controlling intracellular Cu homeostasis to avoid toxicity and ensure delivery of Cu for cbb3-Cox assembly in R. capsulatus. Intriguingly, studies in R. sphaeroides indicated that CcoA is dedicated solely to the biogenesis of cbb3-Cox, but not required for the biogenesis of aa3-Cox. Thus, two distinct Cu delivery pathways operate for Cu insertion into two similar cuproenzymes [110].
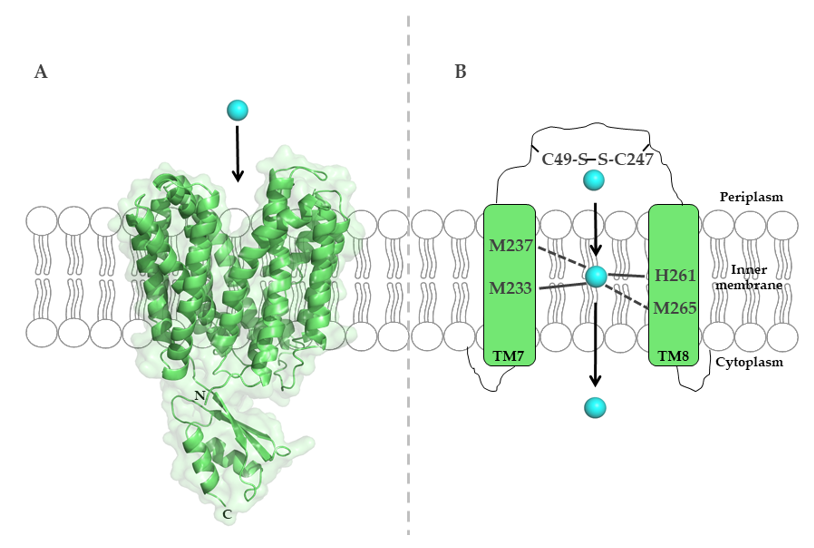
To uncover the molecular mechanisms of Cu binding to CcoA, sequence alignments of R. capsulatus CcoA with other proteobacterial homologues identified two well conserved motifs, M233xxxM237 and H261xxxM265 (R. capsulatus numbering) [113], predicted within transmembrane domains (TMDs) 7 and 8. Mutations within both putative metal-binding sites block Cu import and cbb3-Cox assembly [113] (Figure 4).
Figure 4. The major-facilitator-superfamily (MFS)-like Cu importer CcoA from R. capsulatus. (A) Structure of E. coli YajR (PDB 3WDO), which is homologous to R. capsulatus CcoA, but contains a large cytosolic domain, which is absent in CcoA. (B) Schematic representation of the metal-binding site in R. capsulatus CcoA, which is composed of methionine and histidine residues in transmembrane helices 7 and 8. Cysteine residues in the periplasmic loop likely contribute to Cu transport via CcoA.
A comparative genomic analysis of orthologues in α-proteobacterial species showed that CcoA-like homologues are widespread among these organisms, and frequently co-occur with Cox enzymes [110,114][110][114]. CalT family members also include the RfnT proteins [114], earlier suggested to transport riboflavin [115[115][116],116], but now shown to transport Cu [114]. However, the RfnT-like proteins are unable to restore the cbb3-Cox defects in R. capsulatus ccoA mutants. The lack of functional complementation between CcoA and RfnT-like proteins suggests that even though Cu may be imported via those proteins, its further use is determined by the functional interaction between the Cu importer and the down-stream Cu-binding proteins. Future studies are required to characterize the Cu uptake mechanism and specificity of the CalT family proteins in proteobacteria.
- Argüello, J.M.; Raimunda, D.; Padilla-Benavides, T. Mechanisms of copper homeostasis in bacteria. Cell. Infect. Microbiol. 2013, 3, 73, doi:10.3389/fcimb.2013.00073.
- Giachino, A.; Waldron, K.J. Copper tolerance in bacteria requires the activation of multiple accessory pathways. Microbiol. 2020, doi:10.1111/mmi.14522.
- Ibrahim, Z.; Petrusan, A.J.; Hooke, P.; Hinsa-Leasure, S.M. Reduction of bacterial burden by copper alloys on high-touch athletic center surfaces. J. Infect. Control 2018, 46, 197–201, doi:10.1016/j.ajic.2017.08.028.
- Besold, A.N.; Culbertson, E.M.; Culotta, V.C. The Yin and Yang of copper during infection. Biol. Inorg. Chem. 2016, 21, 137–144, doi:10.1007/s00775-016-1335-1.
- Ladomersky, E.; Khan, A.; Shanbhag, V.; Cavet, J.S.; Chan, J.; Weisman, G.A.; Petris, M.J. Host and Pathogen Copper-Transporting P-Type ATPases Function Antagonistically during Salmonella Infection. Immun. 2017, 85, doi:10.1128/iai.00351-17.
- Antoine, R.; Rivera-Millot, A.; Roy, G.; Jacob-Dubuisson, F. Relationships Between Copper-Related Proteomes and Lifestyles in β Proteobacteria. Microbiol. 2019, 10, 2217, doi:10.3389/fmicb.2019.02217.
- Ward, S.K.; Hoye, E.A.; Talaat, A.M. The global responses of Mycobacterium tuberculosis to physiological levels of copper. Bacteriol. 2008, 190, 2939–2946, doi:10.1128/jb.01847-07.
- Ward, S.K.; Abomoelak, B.; Hoye, E.A.; Steinberg, H.; Talaat, A.M. CtpV: A putative copper exporter required for full virulence of Mycobacterium tuberculosis. Microbiol. 2010, 77, 1096–1110, doi:10.1111/j.1365-2958.2010.07273.x.
- Wolschendorf, F.; Ackart, D.; Shrestha, T.B.; Hascall-Dove, L.; Nolan, S.; Lamichhane, G.; Wang, Y.; Bossmann, S.H.; Basaraba, R.J.; Niederweis, M. Copper resistance is essential for virulence of Mycobacterium tuberculosis. Natl. Acad. Sci. USA 2011, 108, 1621–1626, doi:10.1073/pnas.1009261108.
- Price, E.E.; Boyd, J.M. Genetic Regulation of Metal Ion Homeostasis in Staphylococcus aureus. Trends Microbiol. 2020, 10.1016/j.tim.2020.04.004, doi:10.1016/j.tim.2020.04.004.
- Begg, S.L. The role of metal ions in the virulence and viability of bacterial pathogens. Soc. Trans. 2019, 47, 77–87, doi:10.1042/bst20180275.
- Li, C.; Li, Y.; Ding, C. The Role of Copper Homeostasis at the Host-Pathogen Axis: From Bacteria to Fungi. J. Mol. Sci. 2019, 20, 175, doi:10.3390/ijms20010175.
- Dupont, C.L.; Grass, G.; Rensing, C. Copper toxicity and the origin of bacterial resistance--new insights and applications. Integr. Biometal Sci. 2011, 3, 1109–1118, doi:10.1039/c1mt00107h.
- Shi, X.; Darwin, K.H. Copper homeostasis in Mycobacterium tuberculosis. Integr. Biometal Sci. 2015, 7, 929–934, doi:10.1039/c4mt00305e.
- Braymer, J.J.; Giedroc, D.P. Recent developments in copper and zinc homeostasis in bacterial pathogens. Opin. Chem. Biol. 2014, 19, 59–66, doi:10.1016/j.cbpa.2013.12.021.
- Foster, A.W.; Osman, D.; Robinson, N.J. Metal preferences and metallation. Biol. Chem. 2014, 289, 28095–28103, doi:10.1074/jbc.R114.588145.
- Gaetke, L.M.; Chow, C.K. Copper toxicity, oxidative stress, and antioxidant nutrients. Toxicology 2003, 189, 147–163.
- Stewart, L.J.; Thaqi, D.; Kobe, B.; McEwan, A.G.; Waldron, K.J.; Djoko, K.Y. Handling of nutrient copper in the bacterial envelope. Integr. Biometal Sci. 2019, 11, 50–63, doi:10.1039/C8MT00218E.
- Durand, A.; Azzouzi, A.; Bourbon, M.L.; Steunou, A.S.; Liotenberg, S.; Maeshima, A.; Astier, C.; Argentini, M.; Saito, S.; Ouchane, S. c-Type Cytochrome Assembly Is a Key Target of Copper Toxicity within the Bacterial Periplasm. mBio 2015, 6, e01007–e01015.
- Steunou, A.S.; Durand, A.; Bourbon, M.L.; Babot, M.; Tambosi, R.; Liotenberg, S.; Ouchane, S. Cadmium and Copper Cross-Tolerance. Cu+ Alleviates Cd2+ Toxicity, and Both Cations Target Heme and Chlorophyll Biosynthesis Pathway in Rubrivivax gelatinosus. Microbiol. 2020, 11, 893, doi:10.3389/fmicb.2020.00893.
- Hiniker, A.; Collet, J.F.; Bardwell, J.C. Copper stress causes an in vivo requirement for the Escherichia coli disulfide isomerase DsbC. Biol. Chem. 2005, 280, 33785–33791, doi:10.1074/jbc.M505742200.
- Decker, H.; Terwilliger, N. Cops and robbers: Putative evolution of copper oxygen-binding proteins. Exp. Biol. 2000, 203, 1777–1782.
- Ekici, S.; Yang, H.; Koch, H.-G.; Daldal, F. Novel transporter required for biogenesis of cbb3-type cytochrome c oxidase in Rhodobacter capsulatus. mBio 2012, 3, doi:10.1128/mBio.00293-11.
- Banci, L.; Bertini, I.; Cantini, F.; Ciofi-Baffoni, S. Cellular copper distribution: A mechanistic systems biology approach. Mol. Life Sci. CMLS 2010, 67, 2563–2589, doi:10.1007/s00018-010-0330-x.
- Rensing, C.; McDevitt, S.F. The copper metallome in prokaryotic cells. Ions. Life Sci. 2013, 12, 417–450, doi:10.1007/978-94-007-5561-1_12.
- Aguirre, G.; Pilon, M. Copper Delivery to Chloroplast Proteins and its Regulation. Plant Sci. 2015, 6, 1250, doi:10.3389/fpls.2015.01250.
- Canonica, F.; Klose, D.; Ledermann, R.; Sauer, M.M.; Abicht, H.K.; Quade, N.; Gossert, A.D.; Chesnov, S.; Fischer, H.M.; Jeschke, G.; et al. Structural basis and mechanism for metallochaperone-assisted assembly of the Cu(A) center in cytochrome oxidase. Adv. 2019, 5, eaaw8478, doi:10.1126/sciadv.aaw8478.
- Ekici, S.; Pawlik, G.; Lohmeyer, E.; Koch, H.-G.; Daldal, F. Biogenesis of cbb(3)-type cytochrome c oxidase in Rhodobacter capsulatus. Biophys. Acta 2012, 1817, 898–910, doi:10.1016/j.bbabio.2011.10.011.
- Kudva, R.; Denks, K.; Kuhn, P.; Vogt, A.; Muller, M.; Koch, H.G. Protein translocation across the inner membrane of Gram-negative bacteria: The Sec and Tat dependent protein transport pathways. Microbiol. 2013, 164, 505–534, doi:10.1016/j.resmic.2013.03.016.
- Stolle, P.; Hou, B.; Brüser, T. The Tat Substrate CueO Is Transported in an Incomplete Folding State. Biol. Chem. 2016, 291, 13520–13528, doi:10.1074/jbc.M116.729103.
- Koch, H.G.; Winterstein, C.; Saribas, A.S.; Alben, J.O.; Daldal, F. Roles of the ccoGHIS gene products in the biogenesis of the cbb(3)-type cytochrome c oxidase. Mol. Biol. 2000, 297, 49–65, doi:10.1006/jmbi.2000.3555.
- Kulajta, C.; Thumfart, J.O.; Haid, S.; Daldal, F.; Koch, H.G. Multi-step assembly pathway of the cbb3-type cytochrome c oxidase complex. Mol. Biol. 2006, 355, 989–1004, doi:10.1016/j.jmb.2005.11.039.
- Khalfaoui-Hassani, B.; Verissimo, A.F.; Shroff, N.P.; Ekici, S.; Trasnea, P.-I.; Utz, M.; Koch, H.-G.; Daldal, F. Biogenesis of Cytochrome c Complexes: From Insertion of Redox Cofactors to Assembly of Different Subunits. In Cytochrome Complexes: Evolution, Structures, Energy Transduction, and Signaling; Cramer, W.A., Kallas, T., Eds.; Springer Netherlands: Dordrecht, The Netherlands, 2016; pp. 527–554.
- Richter, O.M.; Ludwig, B. Cytochrome c oxidase—Structure, function, and physiology of a redox-driven molecular machine. Physiol. Biochem. Pharmacol. 2003, 147, 47–74, doi:10.1007/s10254-003-0006-0.
- Ross, M.O.; Rosenzweig, A.C. A tale of two methane monooxygenases. Biol. Inorg. Chem. JBIC Publ. Soc. Biol. Inorg. Chem. 2017, 22, 307–319, doi:10.1007/s00775-016-1419-y.
- Roberts, S.A.; Weichsel, A.; Grass, G.; Thakali, K.; Hazzard, J.T.; Tollin, G.; Rensing, C.; Montfort, W.R. Crystal structure and electron transfer kinetics of CueO, a multicopper oxidase required for copper homeostasis in Escherichia coli. Natl. Acad. Sci. USA 2002, 99, 2766–2771, doi:10.1073/pnas.052710499.
- Horrell, S.; Kekilli, D.; Strange, R.W.; Hough, M.A. Recent structural insights into the function of copper nitrite reductases. Integr. Biometal Sci. 2017, 9, 1470–1482, doi:10.1039/c7mt00146k.
- Palm-Espling, M.E.; Niemiec, M.S.; Wittung-Stafshede, P. Role of metal in folding and stability of copper proteins in vitro. Biophys. Acta 2012, 1823, 1594–1603, doi:10.1016/j.bbamcr.2012.01.013.
- Fenlon, L.A.; Slauch, J.M. Cytoplasmic Copper Detoxification in Salmonella Can Contribute to SodC Metalation but Is Dispensable during Systemic Infection. Bacteriol. 2017, 199, e00437–e00417.
- Zhang, L.; Wüst, A.; Prasser, B.; Müller, C.; Einsle, O. Functional assembly of nitrous oxide reductase provides insights into copper site maturation. Natl. Acad. Sci. USA 2019, 116, 12822–12827, doi:10.1073/pnas.1903819116.
- Buffoni, F.; Ignesti, G. The copper-containing amine oxidases: Biochemical aspects and functional role. Genet. Metab. 2000, 71, 559–564, doi:10.1006/mgme.2000.3098.
- Hermann, B.; Kern, M.; La Pietra, L.; Simon, J.; Einsle, O. The octahaem MccA is a haem c-copper sulfite reductase. Nature 2015, 520, 706–709, doi:10.1038/nature14109.
- Kanteev, M.; Goldfeder, M.; Fishman, A. Structure-function correlations in tyrosinases. Protein Sci. Publ. Protein Soc. 2015, 24, 1360–1369, doi:10.1002/pro.2734.
- McGuirl, M.A.; Bollinger, J.A.; Cosper, N.; Scott, R.A.; Dooley, D.M. Expression, purification, and characterization of NosL, a novel Cu(I) protein of the nitrous oxide reductase (nos) gene cluster. Biol. Inorg. Chem. JBIC Publ. Soc. Biol. Inorg. Chem. 2001, 6, 189–195, doi:10.1007/s007750000190.
- Bennett, S.P.; Soriano-Laguna, M.J.; Bradley, J.M.; Svistunenko, D.A.; Richardson, D.J.; Gates, A.J.; Le Brun, N.E. NosL is a dedicated copper chaperone for assembly of the Cu(Z) center of nitrous oxide reductase. Sci. 2019, 10, 4985–4993, doi:10.1039/c9sc01053j.
- Lohmeyer, E.; Schröder, S.; Pawlik, G.; Trasnea, P.-I.; Peters, A.; Daldal, F.; Koch, H.-G. The ScoI homologue SenC is a copper binding protein that interacts directly with the cbb3-type cytochrome oxidase in Rhodobacter capsulatus. Biophys. Acta 2012, 1817, 2005–2015, doi:10.1016/j.bbabio.2012.06.621.
- Trasnea, P.I.; Andrei, A.; Marckmann, D.; Utz, M.; Khalfaoui-Hassani, B.; Selamoglu, N.; Daldal, F.; Koch, H.G. A Copper Relay System Involving Two Periplasmic Chaperones Drives cbb3-Type Cytochrome c Oxidase Biogenesis in Rhodobacter capsulatus. ACS Chem. Biol. 2018, 13, 1388–1396, doi:10.1021/acschembio.8b00293.
- Serventi, F.; Youard, Z.A.; Murset, V.; Huwiler, S.; Buhler, D.; Richter, M.; Luchsinger, R.; Fischer, H.M.; Brogioli, R.; Niederer, M.; et al. Copper starvation-inducible protein for cytochrome oxidase biogenesis in Bradyrhizobium japonicum. Biol. Chem. 2012, 287, 38812–38823, doi:10.1074/jbc.M112.406173.
- Canonica, F.; Hennecke, H.; Glockshuber, R. Biochemical pathway for the biosynthesis of the Cu(A) center in bacterial cytochrome c oxidase. FEBS Lett. 2019, 593, 2977–2989, doi:10.1002/1873-3468.13587.
- Abicht, H.K.; Scharer, M.A.; Quade, N.; Ledermann, R.; Mohorko, E.; Capitani, G.; Hennecke, H.; Glockshuber, R. How periplasmic thioredoxin TlpA reduces bacterial copper chaperone ScoI and cytochrome oxidase subunit II (CoxB) prior to metallation. Biol. Chem. 2014, 289, 32431–32444, doi:10.1074/jbc.M114.607127.
- Banci, L.; Bertini, I.; Ciofi-Baffoni, S.; Katsari, E.; Katsaros, N.; Kubicek, K.; Mangani, S. A copper(I) protein possibly involved in the assembly of CuA center of bacterial cytochrome c oxidase. Natl. Acad. Sci. USA 2005, 102, 3994–3999, doi:10.1073/pnas.0406150102.
- Trasnea, P.-I.; Utz, M.; Khalfaoui-Hassani, B.; Lagies, S.; Daldal, F.; Koch, H.-G. Cooperation between two periplasmic copper chaperones is required for full activity of the cbb3-type cytochrome c oxidase and copper homeostasis in Rhodobacter capsulatus. Microbiol. 2016, 100, 345–361, doi:10.1111/mmi.13321.
- Thompson, A.K.; Gray, J.; Liu, A.; Hosler, J.P. The roles of Rhodobacter sphaeroides copper chaperones PCu(A)C and Sco (PrrC) in the assembly of the copper centers of the aa(3)-type and the cbb(3)-type cytochrome c oxidases. Biophys. Acta 2012, 1817, 955–964, doi:10.1016/j.bbabio.2012.01.003.
- Rubino, J.T.; Franz, K.J. Coordination chemistry of copper proteins: How nature handles a toxic cargo for essential function. Inorg. Biochem. 2012, 107, 129–143.
- Arguello, J.M.; Gonzalez-Guerrero, M.; Raimunda, D. Bacterial transition metal P(1B)-ATPases: Transport mechanism and roles in virulence. Biochemistry 2011, 50, 9940–9949, doi:10.1021/bi201418k.
- Mattle, D.; Zhang, L.; Sitsel, O.; Pedersen, L.T.; Moncelli, M.R.; Tadini-Buoninsegni, F.; Gourdon, P.; Rees, D.C.; Nissen, P.; Meloni, G. A sulfur-based transport pathway in Cu+-ATPases. EMBO Rep. 2015, 16, 728–740, doi:10.15252/embr.201439927.
- Gourdon, P.; Liu, X.Y.; Skjorringe, T.; Morth, J.P.; Moller, L.B.; Pedersen, B.P.; Nissen, P. Crystal structure of a copper-transporting PIB-type ATPase. Nature 2011, 475, 59–64, doi:10.1038/nature10191.
- Osman, D.; Cavet, J.S. Copper homeostasis in bacteria. Appl. Microbiol. 2008, 65, 217–247, doi:10.1016/s0065-2164(08)00608-4.
- Schweigel-Röntgen, M. The families of zinc (SLC30 and SLC39) and copper (SLC31) transporters. Top. Membr. 2014, 73, 321–355, doi:10.1016/b978-0-12-800223-0.00009-8.
- Dancis, A.; Yuan, D.S.; Haile, D.; Askwith, C.; Eide, D.; Moehle, C.; Kaplan, J.; Klausner, R.D. Molecular characterization of a copper transport protein in S. cerevisiae: An unexpected role for copper in iron transport. Cell 1994, 76, 393–402, doi:10.1016/0092-8674(94)90345-X.
- Öhrvik, H.; Thiele, D.J. The role of Ctr1 and Ctr2 in mammalian copper homeostasis and platinum-based chemotherapy. Trace Elem. Med. Biol. 2015, 31, 178–182, doi:10.1016/j.jtemb.2014.03.006.
- Petris, M.J. The SLC31 (Ctr) copper transporter family. Pflügers Arch. 2004, 447, 752–755, doi:10.1007/s00424-003-1092-1.
- Ren, F.; Logeman, B.L.; Zhang, X.; Liu, Y.; Thiele, D.J.; Yuan, P. X-ray structures of the high-affinity copper transporter Ctr1. Commun. 2019, 10, 1386, doi:10.1038/s41467-019-09376-7.
- Dyla, M.; Kjærgaard, M.; Poulsen, H.; Nissen, P. Structure and Mechanism of P-Type ATPase Ion Pumps. Rev. Biochem. 2020, 89, 583–603, doi:10.1146/annurev-biochem-010611-112801.
- Kanamaru, K.; Kashiwagi, S.; Mizuno, T. A copper-transporting P-type ATPase found in the thylakoid membrane of the cyanobacterium Synechococcus species PCC7942. Microbiol. 1994, 13, 369–377, doi:10.1111/j.1365-2958.1994.tb00430.x.
- Tottey, S.; Rich, P.R.; Rondet, S.A.M.; Robinson, N.J. Two Menkes-type ATPases Supply Copper for Photosynthesis inSynechocystis PCC 6803. Biol. Chem. 2001, 276, 19999–20004.
- Solioz, M. Chapter 11—Copper Disposition in Bacteria. In Clinical and Translational Perspectives on WILSON DISEASE; Kerkar, N., Roberts, E.A., Eds.; Academic Press: Cambridge, MA, USA, 2019; pp. 101–113.
- Solioz, M.; Stoyanov, J.V. Copper homeostasis in Enterococcus hirae. FEMS Microbiol. Rev. 2003, 27, 183–195.
- Badarau, A.; Dennison, C. Thermodynamics of copper and zinc distribution in the cyanobacterium Synechocystis PCC 6803. Natl. Acad. Sci. USA 2011, 108, 13007–13012, doi:10.1073/pnas.1101448108.
- Raimunda, D.; Gonzalez-Guerrero, M.; Leeber, B.W., 3rd; Arguello, J.M. The transport mechanism of bacterial Cu+-ATPases: Distinct efflux rates adapted to different function. Biometals 2011, 24, 467–475, doi:10.1007/s10534-010-9404-3.
- Robinson, N.J.; Winge, D.R. Copper metallochaperones. Rev. Biochem. 2010, 79, 537–562, doi:10.1146/annurev-biochem-030409-143539.
- Teitzel, G.M.; Geddie, A.; De Long, S.K.; Kirisits, M.J.; Whiteley, M.; Parsek, M.R. Survival and Growth in the Presence of Elevated Copper: Transcriptional Profiling of Copper-Stressed Pseudomonas aeruginosa. Bacteriol. 2006, 188, 7242, doi:10.1128/JB.00837-06.
- Almárcegui, R.J.; Navarro, C.A.; Paradela, A.; Albar, J.P.; von Bernath, D.; Jerez, C.A. New Copper Resistance Determinants in the Extremophile Acidithiobacillus ferrooxidans: A Quantitative Proteomic Analysis. Proteome Res. 2014, 13, 946–960, doi:10.1021/pr4009833.
- Selamoglu, N.; Önder, Ö.; Öztürk, Y.; Khalfaoui-Hassani, B.; Blaby-Haas, C.E.; Garcia, B.A.; Koch, H.-G.; Daldal, F. Comparative differential cuproproteomes of Rhodobacter capsulatus reveal novel copper homeostasis related proteins. Metallomics 2020, 12, 572–591, doi:10.1039/C9MT00314B.
- Lutkenhaus, J.F. Role of a major outer membrane protein in Escherichia coli. Bacteriol. 1977, 131, 631–637.
- Li, X.Z.; Nikaido, H.; Williams, K.E. Silver-resistant mutants of Escherichia coli display active efflux of Ag+ and are deficient in porins. Bacteriol. 1997, 179, 6127–6132, doi:10.1128/jb.179.19.6127-6132.1997.
- Niederweis, M.; Danilchanka, O.; Huff, J.; Hoffmann, C.; Engelhardt, H. Mycobacterial outer membranes: in search of proteins. Trends Microbiol. 2010, 18, 109–116, doi:10.1016/j.tim.2009.12.005.
- Alderwick, L.J.; Harrison, J.; Lloyd, G.S.; Birch, H.L. The Mycobacterial Cell Wall--Peptidoglycan and Arabinogalactan. Cold Spring Harbor Perspect. Med. 2015, 5, a021113, doi:10.1101/cshperspect.a021113.
- Speer, A.; Rowland, J.L.; Haeili, M.; Niederweis, M.; Wolschendorf, F. Porins Increase Copper Susceptibility of Mycobacterium tuberculosis. Bacteriol. 2013, 195, 5133, doi:10.1128/JB.00763-13.
- Faller, M.; Niederweis, M.; Schulz, G.E. The Structure of a Mycobacterial Outer-Membrane Channel. Science 2004, 303, 1189, doi:10.1126/science.1094114.
- Niederweis, M.; Ehrt, S.; Heinz, C.; Klöcker, U.; Karosi, S.; Swiderek, K.M.; Riley, L.W.; Benz, R. Cloning of the mspA gene encoding a porin from Mycobacterium smegmatis. Microbiol. 1999, 33, 933–945, doi:10.1046/j.1365-2958.1999.01472.x.
- Shah, S.; Dalecki, A.G.; Malalasekera, A.P.; Crawford, C.L.; Michalek, S.M.; Kutsch, O.; Sun, J.; Bossmann, S.H.; Wolschendorf, F. 8-Hydroxyquinolines Are Boosting Agents of Copper-Related Toxicity in Mycobacterium tuberculosis. Agents Chemother. 2016, 60, 5765, doi:10.1128/AAC.00325-16.
- Carreira, C.; Pauleta, S.R.; Moura, I. The catalytic cycle of nitrous oxide reductase — The enzyme that catalyzes the last step of denitrification. Inorg. Biochem. 2017, 177, 423–434, doi:10.1016/j.jinorgbio.2017.09.007.
- Pomowski, A.; Zumft, W.G.; Kroneck, P.M.H.; Einsle, O. N2O binding at a [4Cu:2S] copper–sulphur cluster in nitrous oxide reductase. Nature 2011, 477, 234–237, doi:10.1038/nature10332.
- Mokhele, K.; Tang, Y.J.; Clark, M.A.; Ingraham, J.L. A Pseudomonas stutzeri outer membrane protein inserts copper into N2O reductase. Bacteriol. 1987, 169, 5721, doi:10.1128/jb.169.12.5721-5726.1987.
- Lee, H.S.; Abdelal, A.H.; Clark, M.A.; Ingraham, J.L. Molecular characterization of nosA, a Pseudomonas stutzeri gene encoding an outer membrane protein required to make copper-containing N2O reductase. Bacteriol. 1991, 173, 5406, doi:10.1128/jb.173.17.5406-5413.1991.
- Schalk, I.J.; Mislin, G.L.; Brillet, K. Structure, function and binding selectivity and stereoselectivity of siderophore-iron outer membrane transporters. Top. Membr. 2012, 69, 37–66, doi:10.1016/b978-0-12-394390-3.00002-1.
- Schalk, I.J.; Hannauer, M.; Braud, A. New roles for bacterial siderophores in metal transport and tolerance. Microbiol. 2011, 13, 2844–2854, doi:10.1111/j.1462-2920.2011.02556.x.
- Schalk, I.J.; Cunrath, O. An overview of the biological metal uptake pathways in Pseudomonas aeruginosa. Microbiol. 2016, 18, 3227–3246, doi:10.1111/1462-2920.13525.
- Rechnitzer, H.; Brzuszkiewicz, E.; Strittmatter, A.; Liesegang, H.; Lysnyansky, I.; Daniel, R.; Gottschalk, G.; Rottem, S. Genomic features and insights into the biology of Mycoplasma fermentans. Microbiology 2011, 157, 760–773, doi:10.1099/mic.0.043208-0.
- Lee, H.S.; Hancock, R.E.; Ingraham, J.L. Properties of a Pseudomonas stutzeri outer membrane channel-forming protein (NosA) required for production of copper-containing N2O reductase. Bacteriol. 1989, 171, 2096, doi:10.1128/jb.171.4.2096-2100.1989.
- Wunsch, P.; Herb, M.; Wieland, H.; Schiek, U.M.; Zumft, W.G. Requirements for CuA and Cu-S Center Assembly of Nitrous Oxide Reductase Deduced from Complete Periplasmic Enzyme Maturation in the Nondenitrifier Pseudomonas putida. Bacteriol. 2003, 185, 887, doi:10.1128/JB.185.3.887-896.2003.
- Chillappagari, S.; Miethke, M.; Trip, H.; Kuipers, O.P.; Marahiel, M.A. Copper acquisition is mediated by YcnJ and regulated by YcnK and CsoR in Bacillus subtilis. Bacteriol. 2009, 191, 2362–2370, doi:10.1128/jb.01616-08.
- Yoneyama, H.; Nakae, T. Protein C (OprC) of the outer membrane of Pseudomonas aeruginosa is a copper-regulated channel protein. Microbiology 1996, 142, 2137–2144, doi:1099/13500872-142-8-2137.
- Quintana, J.; Novoa-Aponte, L.; Argüello, J.M. Copper homeostasis networks in the bacterium Pseudomonas aeruginosa. Biol. Chem. 2017, 292, 15691–15704, doi:10.1074/jbc.M117.804492.
- Han, Y.; Wang, T.; Chen, G.; Pu, Q.; Liu, Q.; Zhang, Y.; Xu, L.; Wu, M.; Liang, H. A Pseudomonas aeruginosa type VI secretion system regulated by CueR facilitates copper acquisition. PLoS Pathog. 2019, 15, e1008198-e1008198, doi:10.1371/journal.ppat.1008198.
- Bhamidimarri, S.P.; Young, T.R.; Shanmugam, M.; Soderholm, S.; Belzunces, B.; Baslé, A.; Skylaris, C.; Bumann, D.; Khalid, S.; van den Berg, B. Acquisition of ionic copper by a bacterial outer membrane protein. bioRxiv 2020, doi:10.1101/2020.06.04.134395.
- Mermod, M.; Magnani, D.; Solioz, M.; Stoyanov, J.V. The copper-inducible ComR (YcfQ) repressor regulates expression of ComC (YcfR), which affects copper permeability of the outer membrane of Escherichia coli. Biometals 2012, 25, 33–43, doi:10.1007/s10534-011-9510-x.
- Egler, M.; Grosse, C.; Grass, G.; Nies, D.H. Role of the Extracytoplasmic Function Protein Family Sigma Factor RpoE in Metal Resistance of Escherichia coli. Bacteriol. 2005, 187, 2297, doi:10.1128/JB.187.7.2297-2307.2005.
- Liu, W.; Wang, Y.; Jing, C. Transcriptome analysis of silver, palladium, and selenium stresses in Pantoea sp. IMH. Chemosphere 2018, 208, 50–58, doi:10.1016/j.chemosphere.2018.05.169.
- Zheng, M.; Wang, X.; Templeton, L.J.; Smulski, D.R.; LaRossa, R.A.; Storz, G. DNA microarray-mediated transcriptional profiling of the Escherichia coli response to hydrogen peroxide. Bacteriol. 2001, 183, 4562–4570, doi:10.1128/jb.183.15.4562-4570.2001.
- Richmond, C.S.; Glasner, J.D.; Mau, R.; Jin, H.; Blattner, F.R. Genome-wide expression profiling in Escherichia coli K-12. Nucleic Acids Res. 1999, 27, 3821–3835, doi:10.1093/nar/27.19.3821.
- Dennison, C.; David, S.; Lee, J. Bacterial copper storage proteins. Biol. Chem. 2018, 293, 4616–4627.
- Pittman, M.S.; Robinson, H.C.; Poole, R.K. A bacterial glutathione transporter (Escherichia coli CydDC) exports reductant to the periplasm. Biol. Chem. 2005, 280, 32254–32261, doi:10.1074/jbc.M503075200.
- Padilla-Benavides, T.; George Thompson, A.M.; McEvoy, M.M.; Arguello, J.M. Mechanism of ATPase-mediated Cu+ export and delivery to periplasmic chaperones: the interaction of Escherichia coli CopA and CusF. Biol. Chem. 2014, 289, 20492–20501, doi:10.1074/jbc.M114.577668.
- Dennison, C. The Coordination Chemistry of Copper Uptake and Storage for Methane Oxidation. Chemistry 2019, 25, 74–86, doi:10.1002/chem.201803444.
- Lee, J.; Dennison, C. Cytosolic Copper Binding by a Bacterial Storage Protein and Interplay with Copper Efflux. J. Mol. Sci. 2019, 20, 4144, doi:10.3390/ijms20174144.
- Quistgaard, E.M.; Löw, C.; Guettou, F.; Nordlund, P. Understanding transport by the major facilitator superfamily (MFS): Structures pave the way. Rev. Mol. Cell Biol. 2016, 17, 123–132, doi:10.1038/nrm.2015.25.
- Ekici, S.; Turkarslan, S.; Pawlik, G.; Dancis, A.; Baliga, N.S.; Koch, H.-G.; Daldal, F. Intracytoplasmic copper homeostasis controls cytochrome c oxidase production. mBio 2014, 5, e01055–e01013, doi:10.1128/mBio.01055-13.
- Khalfaoui-Hassani, B.; Wu, H.; Blaby-Haas, C.E.; Zhang, Y.; Sandri, F.; Verissimo, A.F.; Koch, H.G.; Daldal, F. Widespread Distribution and Functional Specificity of the Copper Importer CcoA: Distinct Cu Uptake Routes for Bacterial Cytochrome c Oxidases. mBio 2018, 9.
- Beaudoin, J.; Ekici, S.; Daldal, F.; Ait-Mohand, S.; Guérin, B.; Labbé, S. Copper transport and regulation in Schizosaccharomyces pombe. Soc. Trans. 2013, 41, 1679–1686, doi:10.1042/BST2013089.
- Arguello, J.M.; Eren, E.; Gonzalez-Guerrero, M. The structure and function of heavy metal transport P1B-ATPases. Biometals 2007, 20, 233–248, doi:10.1007/s10534-006-9055-6.
- Khalfaoui-Hassani, B.; Verissimo, A.F.; Koch, H.-G.; Daldal, F. Uncovering the Transmembrane Metal Binding Site of the Novel Bacterial Major Facilitator Superfamily-Type Copper Importer CcoA. mBio 2016, 7, doi:10.1128/mBio.01981-15.
- Zhang, Y.; Blaby-Haas, C.E.; Steimle, S.; Verissimo, A.F.; Garcia-Angulo, V.A.; Koch, H.-G.; Daldal, F.; Khalfaoui-Hassani, B. Cu Transport by the Extended Family of CcoA-like Transporters (CalT) in Proteobacteria. Rep. 2019, 9, 1208, doi:10.1038/s41598-018-37988-4.
- Gutiérrez-Preciado, A.; Torres, A.G.; Merino, E.; Bonomi, H.R.; Goldbaum, F.A.; García-Angulo, V.A. Extensive Identification of Bacterial Riboflavin Transporters and Their Distribution across Bacterial Species. PLoS ONE 2015, 10, e0126124, doi:10.1371/journal.pone.0126124.
- Vitreschak, A.G.; Rodionov, D.A.; Mironov, A.A.; Gelfand, M.S. Regulation of riboflavin biosynthesis and transport genes in bacteria by transcriptional and translational attenuation. Nucleic Acids Res. 2002, 30, 3141–3151, doi:10.1093/nar/gkf433.
References
- Argüello, J.M.; Raimunda, D.; Padilla-Benavides, T. Mechanisms of copper homeostasis in bacteria. Cell. Infect. Microbiol. 2013, 3, 73, doi:10.3389/fcimb.2013.00073.
- Giachino, A.; Waldron, K.J. Copper tolerance in bacteria requires the activation of multiple accessory pathways. Microbiol. 2020, doi:10.1111/mmi.14522.
- Ibrahim, Z.; Petrusan, A.J.; Hooke, P.; Hinsa-Leasure, S.M. Reduction of bacterial burden by copper alloys on high-touch athletic center surfaces. J. Infect. Control 2018, 46, 197–201, doi:10.1016/j.ajic.2017.08.028.
- Besold, A.N.; Culbertson, E.M.; Culotta, V.C. The Yin and Yang of copper during infection. Biol. Inorg. Chem. 2016, 21, 137–144, doi:10.1007/s00775-016-1335-1.
- Ladomersky, E.; Khan, A.; Shanbhag, V.; Cavet, J.S.; Chan, J.; Weisman, G.A.; Petris, M.J. Host and Pathogen Copper-Transporting P-Type ATPases Function Antagonistically during Salmonella Infection. Immun. 2017, 85, doi:10.1128/iai.00351-17.
- Antoine, R.; Rivera-Millot, A.; Roy, G.; Jacob-Dubuisson, F. Relationships Between Copper-Related Proteomes and Lifestyles in β Proteobacteria. Microbiol. 2019, 10, 2217, doi:10.3389/fmicb.2019.02217.
- Ward, S.K.; Hoye, E.A.; Talaat, A.M. The global responses of Mycobacterium tuberculosis to physiological levels of copper. Bacteriol. 2008, 190, 2939–2946, doi:10.1128/jb.01847-07.
- Ward, S.K.; Abomoelak, B.; Hoye, E.A.; Steinberg, H.; Talaat, A.M. CtpV: A putative copper exporter required for full virulence of Mycobacterium tuberculosis. Microbiol. 2010, 77, 1096–1110, doi:10.1111/j.1365-2958.2010.07273.x.
- Wolschendorf, F.; Ackart, D.; Shrestha, T.B.; Hascall-Dove, L.; Nolan, S.; Lamichhane, G.; Wang, Y.; Bossmann, S.H.; Basaraba, R.J.; Niederweis, M. Copper resistance is essential for virulence of Mycobacterium tuberculosis. Natl. Acad. Sci. USA 2011, 108, 1621–1626, doi:10.1073/pnas.1009261108.
- Price, E.E.; Boyd, J.M. Genetic Regulation of Metal Ion Homeostasis in Staphylococcus aureus. Trends Microbiol. 2020, 10.1016/j.tim.2020.04.004, doi:10.1016/j.tim.2020.04.004.
- Begg, S.L. The role of metal ions in the virulence and viability of bacterial pathogens. Soc. Trans. 2019, 47, 77–87, doi:10.1042/bst20180275.
- Li, C.; Li, Y.; Ding, C. The Role of Copper Homeostasis at the Host-Pathogen Axis: From Bacteria to Fungi. J. Mol. Sci. 2019, 20, 175, doi:10.3390/ijms20010175.
- Dupont, C.L.; Grass, G.; Rensing, C. Copper toxicity and the origin of bacterial resistance--new insights and applications. Integr. Biometal Sci. 2011, 3, 1109–1118, doi:10.1039/c1mt00107h.
- Shi, X.; Darwin, K.H. Copper homeostasis in Mycobacterium tuberculosis. Integr. Biometal Sci. 2015, 7, 929–934, doi:10.1039/c4mt00305e.
- Braymer, J.J.; Giedroc, D.P. Recent developments in copper and zinc homeostasis in bacterial pathogens. Opin. Chem. Biol. 2014, 19, 59–66, doi:10.1016/j.cbpa.2013.12.021.
- Foster, A.W.; Osman, D.; Robinson, N.J. Metal preferences and metallation. Biol. Chem. 2014, 289, 28095–28103, doi:10.1074/jbc.R114.588145.
- Gaetke, L.M.; Chow, C.K. Copper toxicity, oxidative stress, and antioxidant nutrients. Toxicology 2003, 189, 147–163.
- Stewart, L.J.; Thaqi, D.; Kobe, B.; McEwan, A.G.; Waldron, K.J.; Djoko, K.Y. Handling of nutrient copper in the bacterial envelope. Integr. Biometal Sci. 2019, 11, 50–63, doi:10.1039/C8MT00218E.
- Durand, A.; Azzouzi, A.; Bourbon, M.L.; Steunou, A.S.; Liotenberg, S.; Maeshima, A.; Astier, C.; Argentini, M.; Saito, S.; Ouchane, S. c-Type Cytochrome Assembly Is a Key Target of Copper Toxicity within the Bacterial Periplasm. mBio 2015, 6, e01007–e01015.
- Steunou, A.S.; Durand, A.; Bourbon, M.L.; Babot, M.; Tambosi, R.; Liotenberg, S.; Ouchane, S. Cadmium and Copper Cross-Tolerance. Cu+ Alleviates Cd2+ Toxicity, and Both Cations Target Heme and Chlorophyll Biosynthesis Pathway in Rubrivivax gelatinosus. Microbiol. 2020, 11, 893, doi:10.3389/fmicb.2020.00893.
- Hiniker, A.; Collet, J.F.; Bardwell, J.C. Copper stress causes an in vivo requirement for the Escherichia coli disulfide isomerase DsbC. Biol. Chem. 2005, 280, 33785–33791, doi:10.1074/jbc.M505742200.
- Decker, H.; Terwilliger, N. Cops and robbers: Putative evolution of copper oxygen-binding proteins. Exp. Biol. 2000, 203, 1777–1782.
- Ekici, S.; Yang, H.; Koch, H.-G.; Daldal, F. Novel transporter required for biogenesis of cbb3-type cytochrome c oxidase in Rhodobacter capsulatus. mBio 2012, 3, doi:10.1128/mBio.00293-11.
- Banci, L.; Bertini, I.; Cantini, F.; Ciofi-Baffoni, S. Cellular copper distribution: A mechanistic systems biology approach. Mol. Life Sci. CMLS 2010, 67, 2563–2589, doi:10.1007/s00018-010-0330-x.
- Rensing, C.; McDevitt, S.F. The copper metallome in prokaryotic cells. Ions. Life Sci. 2013, 12, 417–450, doi:10.1007/978-94-007-5561-1_12.
- Aguirre, G.; Pilon, M. Copper Delivery to Chloroplast Proteins and its Regulation. Plant Sci. 2015, 6, 1250, doi:10.3389/fpls.2015.01250.
- Canonica, F.; Klose, D.; Ledermann, R.; Sauer, M.M.; Abicht, H.K.; Quade, N.; Gossert, A.D.; Chesnov, S.; Fischer, H.M.; Jeschke, G.; et al. Structural basis and mechanism for metallochaperone-assisted assembly of the Cu(A) center in cytochrome oxidase. Adv. 2019, 5, eaaw8478, doi:10.1126/sciadv.aaw8478.
- Ekici, S.; Pawlik, G.; Lohmeyer, E.; Koch, H.-G.; Daldal, F. Biogenesis of cbb(3)-type cytochrome c oxidase in Rhodobacter capsulatus. Biophys. Acta 2012, 1817, 898–910, doi:10.1016/j.bbabio.2011.10.011.
- Kudva, R.; Denks, K.; Kuhn, P.; Vogt, A.; Muller, M.; Koch, H.G. Protein translocation across the inner membrane of Gram-negative bacteria: The Sec and Tat dependent protein transport pathways. Microbiol. 2013, 164, 505–534, doi:10.1016/j.resmic.2013.03.016.
- Stolle, P.; Hou, B.; Brüser, T. The Tat Substrate CueO Is Transported in an Incomplete Folding State. Biol. Chem. 2016, 291, 13520–13528, doi:10.1074/jbc.M116.729103.
- Koch, H.G.; Winterstein, C.; Saribas, A.S.; Alben, J.O.; Daldal, F. Roles of the ccoGHIS gene products in the biogenesis of the cbb(3)-type cytochrome c oxidase. Mol. Biol. 2000, 297, 49–65, doi:10.1006/jmbi.2000.3555.
- Kulajta, C.; Thumfart, J.O.; Haid, S.; Daldal, F.; Koch, H.G. Multi-step assembly pathway of the cbb3-type cytochrome c oxidase complex. Mol. Biol. 2006, 355, 989–1004, doi:10.1016/j.jmb.2005.11.039.
- Khalfaoui-Hassani, B.; Verissimo, A.F.; Shroff, N.P.; Ekici, S.; Trasnea, P.-I.; Utz, M.; Koch, H.-G.; Daldal, F. Biogenesis of Cytochrome c Complexes: From Insertion of Redox Cofactors to Assembly of Different Subunits. In Cytochrome Complexes: Evolution, Structures, Energy Transduction, and Signaling; Cramer, W.A., Kallas, T., Eds.; Springer Netherlands: Dordrecht, The Netherlands, 2016; pp. 527–554.
- Richter, O.M.; Ludwig, B. Cytochrome c oxidase—Structure, function, and physiology of a redox-driven molecular machine. Physiol. Biochem. Pharmacol. 2003, 147, 47–74, doi:10.1007/s10254-003-0006-0.
- Ross, M.O.; Rosenzweig, A.C. A tale of two methane monooxygenases. Biol. Inorg. Chem. JBIC Publ. Soc. Biol. Inorg. Chem. 2017, 22, 307–319, doi:10.1007/s00775-016-1419-y.
- Roberts, S.A.; Weichsel, A.; Grass, G.; Thakali, K.; Hazzard, J.T.; Tollin, G.; Rensing, C.; Montfort, W.R. Crystal structure and electron transfer kinetics of CueO, a multicopper oxidase required for copper homeostasis in Escherichia coli. Natl. Acad. Sci. USA 2002, 99, 2766–2771, doi:10.1073/pnas.052710499.
- Horrell, S.; Kekilli, D.; Strange, R.W.; Hough, M.A. Recent structural insights into the function of copper nitrite reductases. Integr. Biometal Sci. 2017, 9, 1470–1482, doi:10.1039/c7mt00146k.
- Palm-Espling, M.E.; Niemiec, M.S.; Wittung-Stafshede, P. Role of metal in folding and stability of copper proteins in vitro. Biophys. Acta 2012, 1823, 1594–1603, doi:10.1016/j.bbamcr.2012.01.013.
- Fenlon, L.A.; Slauch, J.M. Cytoplasmic Copper Detoxification in Salmonella Can Contribute to SodC Metalation but Is Dispensable during Systemic Infection. Bacteriol. 2017, 199, e00437–e00417.
- Zhang, L.; Wüst, A.; Prasser, B.; Müller, C.; Einsle, O. Functional assembly of nitrous oxide reductase provides insights into copper site maturation. Natl. Acad. Sci. USA 2019, 116, 12822–12827, doi:10.1073/pnas.1903819116.
- Buffoni, F.; Ignesti, G. The copper-containing amine oxidases: Biochemical aspects and functional role. Genet. Metab. 2000, 71, 559–564, doi:10.1006/mgme.2000.3098.
- Hermann, B.; Kern, M.; La Pietra, L.; Simon, J.; Einsle, O. The octahaem MccA is a haem c-copper sulfite reductase. Nature 2015, 520, 706–709, doi:10.1038/nature14109.
- Kanteev, M.; Goldfeder, M.; Fishman, A. Structure-function correlations in tyrosinases. Protein Sci. Publ. Protein Soc. 2015, 24, 1360–1369, doi:10.1002/pro.2734.
- McGuirl, M.A.; Bollinger, J.A.; Cosper, N.; Scott, R.A.; Dooley, D.M. Expression, purification, and characterization of NosL, a novel Cu(I) protein of the nitrous oxide reductase (nos) gene cluster. Biol. Inorg. Chem. JBIC Publ. Soc. Biol. Inorg. Chem. 2001, 6, 189–195, doi:10.1007/s007750000190.
- Bennett, S.P.; Soriano-Laguna, M.J.; Bradley, J.M.; Svistunenko, D.A.; Richardson, D.J.; Gates, A.J.; Le Brun, N.E. NosL is a dedicated copper chaperone for assembly of the Cu(Z) center of nitrous oxide reductase. Sci. 2019, 10, 4985–4993, doi:10.1039/c9sc01053j.
- Lohmeyer, E.; Schröder, S.; Pawlik, G.; Trasnea, P.-I.; Peters, A.; Daldal, F.; Koch, H.-G. The ScoI homologue SenC is a copper binding protein that interacts directly with the cbb3-type cytochrome oxidase in Rhodobacter capsulatus. Biophys. Acta 2012, 1817, 2005–2015, doi:10.1016/j.bbabio.2012.06.621.
- Trasnea, P.I.; Andrei, A.; Marckmann, D.; Utz, M.; Khalfaoui-Hassani, B.; Selamoglu, N.; Daldal, F.; Koch, H.G. A Copper Relay System Involving Two Periplasmic Chaperones Drives cbb3-Type Cytochrome c Oxidase Biogenesis in Rhodobacter capsulatus. ACS Chem. Biol. 2018, 13, 1388–1396, doi:10.1021/acschembio.8b00293.
- Serventi, F.; Youard, Z.A.; Murset, V.; Huwiler, S.; Buhler, D.; Richter, M.; Luchsinger, R.; Fischer, H.M.; Brogioli, R.; Niederer, M.; et al. Copper starvation-inducible protein for cytochrome oxidase biogenesis in Bradyrhizobium japonicum. Biol. Chem. 2012, 287, 38812–38823, doi:10.1074/jbc.M112.406173.
- Canonica, F.; Hennecke, H.; Glockshuber, R. Biochemical pathway for the biosynthesis of the Cu(A) center in bacterial cytochrome c oxidase. FEBS Lett. 2019, 593, 2977–2989, doi:10.1002/1873-3468.13587.
- Abicht, H.K.; Scharer, M.A.; Quade, N.; Ledermann, R.; Mohorko, E.; Capitani, G.; Hennecke, H.; Glockshuber, R. How periplasmic thioredoxin TlpA reduces bacterial copper chaperone ScoI and cytochrome oxidase subunit II (CoxB) prior to metallation. Biol. Chem. 2014, 289, 32431–32444, doi:10.1074/jbc.M114.607127.
- Banci, L.; Bertini, I.; Ciofi-Baffoni, S.; Katsari, E.; Katsaros, N.; Kubicek, K.; Mangani, S. A copper(I) protein possibly involved in the assembly of CuA center of bacterial cytochrome c oxidase. Natl. Acad. Sci. USA 2005, 102, 3994–3999, doi:10.1073/pnas.0406150102.
- Trasnea, P.-I.; Utz, M.; Khalfaoui-Hassani, B.; Lagies, S.; Daldal, F.; Koch, H.-G. Cooperation between two periplasmic copper chaperones is required for full activity of the cbb3-type cytochrome c oxidase and copper homeostasis in Rhodobacter capsulatus. Microbiol. 2016, 100, 345–361, doi:10.1111/mmi.13321.
- Thompson, A.K.; Gray, J.; Liu, A.; Hosler, J.P. The roles of Rhodobacter sphaeroides copper chaperones PCu(A)C and Sco (PrrC) in the assembly of the copper centers of the aa(3)-type and the cbb(3)-type cytochrome c oxidases. Biophys. Acta 2012, 1817, 955–964, doi:10.1016/j.bbabio.2012.01.003.
- Rubino, J.T.; Franz, K.J. Coordination chemistry of copper proteins: How nature handles a toxic cargo for essential function. Inorg. Biochem. 2012, 107, 129–143.
- Arguello, J.M.; Gonzalez-Guerrero, M.; Raimunda, D. Bacterial transition metal P(1B)-ATPases: Transport mechanism and roles in virulence. Biochemistry 2011, 50, 9940–9949, doi:10.1021/bi201418k.
- Mattle, D.; Zhang, L.; Sitsel, O.; Pedersen, L.T.; Moncelli, M.R.; Tadini-Buoninsegni, F.; Gourdon, P.; Rees, D.C.; Nissen, P.; Meloni, G. A sulfur-based transport pathway in Cu+-ATPases. EMBO Rep. 2015, 16, 728–740, doi:10.15252/embr.201439927.
- Gourdon, P.; Liu, X.Y.; Skjorringe, T.; Morth, J.P.; Moller, L.B.; Pedersen, B.P.; Nissen, P. Crystal structure of a copper-transporting PIB-type ATPase. Nature 2011, 475, 59–64, doi:10.1038/nature10191.
- Osman, D.; Cavet, J.S. Copper homeostasis in bacteria. Appl. Microbiol. 2008, 65, 217–247, doi:10.1016/s0065-2164(08)00608-4.
- Schweigel-Röntgen, M. The families of zinc (SLC30 and SLC39) and copper (SLC31) transporters. Top. Membr. 2014, 73, 321–355, doi:10.1016/b978-0-12-800223-0.00009-8.
- Dancis, A.; Yuan, D.S.; Haile, D.; Askwith, C.; Eide, D.; Moehle, C.; Kaplan, J.; Klausner, R.D. Molecular characterization of a copper transport protein in S. cerevisiae: An unexpected role for copper in iron transport. Cell 1994, 76, 393–402, doi:10.1016/0092-8674(94)90345-X.
- Öhrvik, H.; Thiele, D.J. The role of Ctr1 and Ctr2 in mammalian copper homeostasis and platinum-based chemotherapy. Trace Elem. Med. Biol. 2015, 31, 178–182, doi:10.1016/j.jtemb.2014.03.006.
- Petris, M.J. The SLC31 (Ctr) copper transporter family. Pflügers Arch. 2004, 447, 752–755, doi:10.1007/s00424-003-1092-1.
- Ren, F.; Logeman, B.L.; Zhang, X.; Liu, Y.; Thiele, D.J.; Yuan, P. X-ray structures of the high-affinity copper transporter Ctr1. Commun. 2019, 10, 1386, doi:10.1038/s41467-019-09376-7.
- Dyla, M.; Kjærgaard, M.; Poulsen, H.; Nissen, P. Structure and Mechanism of P-Type ATPase Ion Pumps. Rev. Biochem. 2020, 89, 583–603, doi:10.1146/annurev-biochem-010611-112801.
- Kanamaru, K.; Kashiwagi, S.; Mizuno, T. A copper-transporting P-type ATPase found in the thylakoid membrane of the cyanobacterium Synechococcus species PCC7942. Microbiol. 1994, 13, 369–377, doi:10.1111/j.1365-2958.1994.tb00430.x.
- Tottey, S.; Rich, P.R.; Rondet, S.A.M.; Robinson, N.J. Two Menkes-type ATPases Supply Copper for Photosynthesis inSynechocystis PCC 6803. Biol. Chem. 2001, 276, 19999–20004.
- Solioz, M. Chapter 11—Copper Disposition in Bacteria. In Clinical and Translational Perspectives on WILSON DISEASE; Kerkar, N., Roberts, E.A., Eds.; Academic Press: Cambridge, MA, USA, 2019; pp. 101–113.
- Solioz, M.; Stoyanov, J.V. Copper homeostasis in Enterococcus hirae. FEMS Microbiol. Rev. 2003, 27, 183–195.
- Badarau, A.; Dennison, C. Thermodynamics of copper and zinc distribution in the cyanobacterium Synechocystis PCC 6803. Natl. Acad. Sci. USA 2011, 108, 13007–13012, doi:10.1073/pnas.1101448108.
- Raimunda, D.; Gonzalez-Guerrero, M.; Leeber, B.W., 3rd; Arguello, J.M. The transport mechanism of bacterial Cu+-ATPases: Distinct efflux rates adapted to different function. Biometals 2011, 24, 467–475, doi:10.1007/s10534-010-9404-3.
- Robinson, N.J.; Winge, D.R. Copper metallochaperones. Rev. Biochem. 2010, 79, 537–562, doi:10.1146/annurev-biochem-030409-143539.
- Teitzel, G.M.; Geddie, A.; De Long, S.K.; Kirisits, M.J.; Whiteley, M.; Parsek, M.R. Survival and Growth in the Presence of Elevated Copper: Transcriptional Profiling of Copper-Stressed Pseudomonas aeruginosa. Bacteriol. 2006, 188, 7242, doi:10.1128/JB.00837-06.
- Almárcegui, R.J.; Navarro, C.A.; Paradela, A.; Albar, J.P.; von Bernath, D.; Jerez, C.A. New Copper Resistance Determinants in the Extremophile Acidithiobacillus ferrooxidans: A Quantitative Proteomic Analysis. Proteome Res. 2014, 13, 946–960, doi:10.1021/pr4009833.
- Selamoglu, N.; Önder, Ö.; Öztürk, Y.; Khalfaoui-Hassani, B.; Blaby-Haas, C.E.; Garcia, B.A.; Koch, H.-G.; Daldal, F. Comparative differential cuproproteomes of Rhodobacter capsulatus reveal novel copper homeostasis related proteins. Metallomics 2020, 12, 572–591, doi:10.1039/C9MT00314B.
- Lutkenhaus, J.F. Role of a major outer membrane protein in Escherichia coli. Bacteriol. 1977, 131, 631–637.
- Li, X.Z.; Nikaido, H.; Williams, K.E. Silver-resistant mutants of Escherichia coli display active efflux of Ag+ and are deficient in porins. Bacteriol. 1997, 179, 6127–6132, doi:10.1128/jb.179.19.6127-6132.1997.
- Niederweis, M.; Danilchanka, O.; Huff, J.; Hoffmann, C.; Engelhardt, H. Mycobacterial outer membranes: in search of proteins. Trends Microbiol. 2010, 18, 109–116, doi:10.1016/j.tim.2009.12.005.
- Alderwick, L.J.; Harrison, J.; Lloyd, G.S.; Birch, H.L. The Mycobacterial Cell Wall--Peptidoglycan and Arabinogalactan. Cold Spring Harbor Perspect. Med. 2015, 5, a021113, doi:10.1101/cshperspect.a021113.
- Speer, A.; Rowland, J.L.; Haeili, M.; Niederweis, M.; Wolschendorf, F. Porins Increase Copper Susceptibility of Mycobacterium tuberculosis. Bacteriol. 2013, 195, 5133, doi:10.1128/JB.00763-13.
- Faller, M.; Niederweis, M.; Schulz, G.E. The Structure of a Mycobacterial Outer-Membrane Channel. Science 2004, 303, 1189, doi:10.1126/science.1094114.
- Niederweis, M.; Ehrt, S.; Heinz, C.; Klöcker, U.; Karosi, S.; Swiderek, K.M.; Riley, L.W.; Benz, R. Cloning of the mspA gene encoding a porin from Mycobacterium smegmatis. Microbiol. 1999, 33, 933–945, doi:10.1046/j.1365-2958.1999.01472.x.
- Shah, S.; Dalecki, A.G.; Malalasekera, A.P.; Crawford, C.L.; Michalek, S.M.; Kutsch, O.; Sun, J.; Bossmann, S.H.; Wolschendorf, F. 8-Hydroxyquinolines Are Boosting Agents of Copper-Related Toxicity in Mycobacterium tuberculosis. Agents Chemother. 2016, 60, 5765, doi:10.1128/AAC.00325-16.
- Carreira, C.; Pauleta, S.R.; Moura, I. The catalytic cycle of nitrous oxide reductase — The enzyme that catalyzes the last step of denitrification. Inorg. Biochem. 2017, 177, 423–434, doi:10.1016/j.jinorgbio.2017.09.007.
- Pomowski, A.; Zumft, W.G.; Kroneck, P.M.H.; Einsle, O. N2O binding at a [4Cu:2S] copper–sulphur cluster in nitrous oxide reductase. Nature 2011, 477, 234–237, doi:10.1038/nature10332.
- Mokhele, K.; Tang, Y.J.; Clark, M.A.; Ingraham, J.L. A Pseudomonas stutzeri outer membrane protein inserts copper into N2O reductase. Bacteriol. 1987, 169, 5721, doi:10.1128/jb.169.12.5721-5726.1987.
- Lee, H.S.; Abdelal, A.H.; Clark, M.A.; Ingraham, J.L. Molecular characterization of nosA, a Pseudomonas stutzeri gene encoding an outer membrane protein required to make copper-containing N2O reductase. Bacteriol. 1991, 173, 5406, doi:10.1128/jb.173.17.5406-5413.1991.
- Schalk, I.J.; Mislin, G.L.; Brillet, K. Structure, function and binding selectivity and stereoselectivity of siderophore-iron outer membrane transporters. Top. Membr. 2012, 69, 37–66, doi:10.1016/b978-0-12-394390-3.00002-1.
- Schalk, I.J.; Hannauer, M.; Braud, A. New roles for bacterial siderophores in metal transport and tolerance. Microbiol. 2011, 13, 2844–2854, doi:10.1111/j.1462-2920.2011.02556.x.
- Schalk, I.J.; Cunrath, O. An overview of the biological metal uptake pathways in Pseudomonas aeruginosa. Microbiol. 2016, 18, 3227–3246, doi:10.1111/1462-2920.13525.
- Rechnitzer, H.; Brzuszkiewicz, E.; Strittmatter, A.; Liesegang, H.; Lysnyansky, I.; Daniel, R.; Gottschalk, G.; Rottem, S. Genomic features and insights into the biology of Mycoplasma fermentans. Microbiology 2011, 157, 760–773, doi:10.1099/mic.0.043208-0.
- Lee, H.S.; Hancock, R.E.; Ingraham, J.L. Properties of a Pseudomonas stutzeri outer membrane channel-forming protein (NosA) required for production of copper-containing N2O reductase. Bacteriol. 1989, 171, 2096, doi:10.1128/jb.171.4.2096-2100.1989.
- Wunsch, P.; Herb, M.; Wieland, H.; Schiek, U.M.; Zumft, W.G. Requirements for CuA and Cu-S Center Assembly of Nitrous Oxide Reductase Deduced from Complete Periplasmic Enzyme Maturation in the Nondenitrifier Pseudomonas putida. Bacteriol. 2003, 185, 887, doi:10.1128/JB.185.3.887-896.2003.
- Chillappagari, S.; Miethke, M.; Trip, H.; Kuipers, O.P.; Marahiel, M.A. Copper acquisition is mediated by YcnJ and regulated by YcnK and CsoR in Bacillus subtilis. Bacteriol. 2009, 191, 2362–2370, doi:10.1128/jb.01616-08.
- Yoneyama, H.; Nakae, T. Protein C (OprC) of the outer membrane of Pseudomonas aeruginosa is a copper-regulated channel protein. Microbiology 1996, 142, 2137–2144, doi:1099/13500872-142-8-2137.
- Quintana, J.; Novoa-Aponte, L.; Argüello, J.M. Copper homeostasis networks in the bacterium Pseudomonas aeruginosa. Biol. Chem. 2017, 292, 15691–15704, doi:10.1074/jbc.M117.804492.
- Han, Y.; Wang, T.; Chen, G.; Pu, Q.; Liu, Q.; Zhang, Y.; Xu, L.; Wu, M.; Liang, H. A Pseudomonas aeruginosa type VI secretion system regulated by CueR facilitates copper acquisition. PLoS Pathog. 2019, 15, e1008198-e1008198, doi:10.1371/journal.ppat.1008198.
- Bhamidimarri, S.P.; Young, T.R.; Shanmugam, M.; Soderholm, S.; Belzunces, B.; Baslé, A.; Skylaris, C.; Bumann, D.; Khalid, S.; van den Berg, B. Acquisition of ionic copper by a bacterial outer membrane protein. bioRxiv 2020, doi:10.1101/2020.06.04.134395.
- Mermod, M.; Magnani, D.; Solioz, M.; Stoyanov, J.V. The copper-inducible ComR (YcfQ) repressor regulates expression of ComC (YcfR), which affects copper permeability of the outer membrane of Escherichia coli. Biometals 2012, 25, 33–43, doi:10.1007/s10534-011-9510-x.
- Egler, M.; Grosse, C.; Grass, G.; Nies, D.H. Role of the Extracytoplasmic Function Protein Family Sigma Factor RpoE in Metal Resistance of Escherichia coli. Bacteriol. 2005, 187, 2297, doi:10.1128/JB.187.7.2297-2307.2005.
- Liu, W.; Wang, Y.; Jing, C. Transcriptome analysis of silver, palladium, and selenium stresses in Pantoea sp. IMH. Chemosphere 2018, 208, 50–58, doi:10.1016/j.chemosphere.2018.05.169.
- Zheng, M.; Wang, X.; Templeton, L.J.; Smulski, D.R.; LaRossa, R.A.; Storz, G. DNA microarray-mediated transcriptional profiling of the Escherichia coli response to hydrogen peroxide. Bacteriol. 2001, 183, 4562–4570, doi:10.1128/jb.183.15.4562-4570.2001.
- Richmond, C.S.; Glasner, J.D.; Mau, R.; Jin, H.; Blattner, F.R. Genome-wide expression profiling in Escherichia coli K-12. Nucleic Acids Res. 1999, 27, 3821–3835, doi:10.1093/nar/27.19.3821.
- Dennison, C.; David, S.; Lee, J. Bacterial copper storage proteins. Biol. Chem. 2018, 293, 4616–4627.
- Pittman, M.S.; Robinson, H.C.; Poole, R.K. A bacterial glutathione transporter (Escherichia coli CydDC) exports reductant to the periplasm. Biol. Chem. 2005, 280, 32254–32261, doi:10.1074/jbc.M503075200.
- Padilla-Benavides, T.; George Thompson, A.M.; McEvoy, M.M.; Arguello, J.M. Mechanism of ATPase-mediated Cu+ export and delivery to periplasmic chaperones: the interaction of Escherichia coli CopA and CusF. Biol. Chem. 2014, 289, 20492–20501, doi:10.1074/jbc.M114.577668.
- Dennison, C. The Coordination Chemistry of Copper Uptake and Storage for Methane Oxidation. Chemistry 2019, 25, 74–86, doi:10.1002/chem.201803444.
- Lee, J.; Dennison, C. Cytosolic Copper Binding by a Bacterial Storage Protein and Interplay with Copper Efflux. J. Mol. Sci. 2019, 20, 4144, doi:10.3390/ijms20174144.
- Quistgaard, E.M.; Löw, C.; Guettou, F.; Nordlund, P. Understanding transport by the major facilitator superfamily (MFS): Structures pave the way. Rev. Mol. Cell Biol. 2016, 17, 123–132, doi:10.1038/nrm.2015.25.
- Ekici, S.; Turkarslan, S.; Pawlik, G.; Dancis, A.; Baliga, N.S.; Koch, H.-G.; Daldal, F. Intracytoplasmic copper homeostasis controls cytochrome c oxidase production. mBio 2014, 5, e01055–e01013, doi:10.1128/mBio.01055-13.
- Khalfaoui-Hassani, B.; Wu, H.; Blaby-Haas, C.E.; Zhang, Y.; Sandri, F.; Verissimo, A.F.; Koch, H.G.; Daldal, F. Widespread Distribution and Functional Specificity of the Copper Importer CcoA: Distinct Cu Uptake Routes for Bacterial Cytochrome c Oxidases. mBio 2018, 9.
- Beaudoin, J.; Ekici, S.; Daldal, F.; Ait-Mohand, S.; Guérin, B.; Labbé, S. Copper transport and regulation in Schizosaccharomyces pombe. Soc. Trans. 2013, 41, 1679–1686, doi:10.1042/BST2013089.
- Arguello, J.M.; Eren, E.; Gonzalez-Guerrero, M. The structure and function of heavy metal transport P1B-ATPases. Biometals 2007, 20, 233–248, doi:10.1007/s10534-006-9055-6.
- Khalfaoui-Hassani, B.; Verissimo, A.F.; Koch, H.-G.; Daldal, F. Uncovering the Transmembrane Metal Binding Site of the Novel Bacterial Major Facilitator Superfamily-Type Copper Importer CcoA. mBio 2016, 7, doi:10.1128/mBio.01981-15.
- Zhang, Y.; Blaby-Haas, C.E.; Steimle, S.; Verissimo, A.F.; Garcia-Angulo, V.A.; Koch, H.-G.; Daldal, F.; Khalfaoui-Hassani, B. Cu Transport by the Extended Family of CcoA-like Transporters (CalT) in Proteobacteria. Rep. 2019, 9, 1208, doi:10.1038/s41598-018-37988-4.
- Gutiérrez-Preciado, A.; Torres, A.G.; Merino, E.; Bonomi, H.R.; Goldbaum, F.A.; García-Angulo, V.A. Extensive Identification of Bacterial Riboflavin Transporters and Their Distribution across Bacterial Species. PLoS ONE 2015, 10, e0126124, doi:10.1371/journal.pone.0126124.
- Vitreschak, A.G.; Rodionov, D.A.; Mironov, A.A.; Gelfand, M.S. Regulation of riboflavin biosynthesis and transport genes in bacteria by transcriptional and translational attenuation. Nucleic Acids Res. 2002, 30, 3141–3151, doi:10.1093/nar/gkf433.