Bioaerosol characterization represents a major challenge for the risk assessment and management of exposed people. One of the most important bioaerosol sources is the organic waste collection and treatment. This work analyzed and discussed the literature with the purpose of investigating the main techniques used nowadays for bioaerosol monitoring during organic waste treatment. The discussion includes an overview on the most effcient sampling, DNA extraction, and analysis methods, including both the cultural and the bio-molecular approach. Generally, an exhaustive biological risk assessment is not applied due to the organic waste heterogeneity, treatment complexity, and unknown aerosolized emission rate. However, the application of bio-molecular methods allows a better bioaerosol characterization, and it is desirable to be associated with standardized cultural methods. Risk assessment for organic waste workers generally includes the evaluation of the potential exposition to pathogens and opportunistic pathogens or to other microorganisms as biomarkers. In most cases,
Bioaerosol characterization represents a major challenge for the risk assessment and
management of exposed people. One of the most important bioaerosol sources is the organic waste
collection and treatment. This work analyzed and discussed the literature with the purpose of
investigating the main techniques used nowadays for bioaerosol monitoring during organic waste
treatment. The discussion includes an overview on the most effcient sampling, DNA extraction,
and analysis methods, including both the cultural and the bio-molecular approach. Generally,
an exhaustive biological risk assessment is not applied due to the organic waste heterogeneity,
treatment complexity, and unknown aerosolized emission rate. However, the application of
bio-molecular methods allows a better bioaerosol characterization, and it is desirable to be associated
with standardized cultural methods. Risk assessment for organic waste workers generally includes
the evaluation of the potential exposition to pathogens and opportunistic pathogens or to other
microorganisms as biomarkers. In most cases,
Saccharopolyspora rectivirgula, Legionella
spp.,
Aspergillus spp., and
spp., and
Mycobacterium spp. are included. Future perspectives are focused on identifying common composting biomarkers, on investigating the causality process between chronic bioaerosol exposure and disease onset, and finally, on defining common exposure limits.
spp. are included. Future perspectives are focused on identifying common
composting biomarkers, on investigating the causality process between chronic bioaerosol exposure
and disease onset, and finally, on defining common exposure limits.
- bioaerosol
- sampling method
- sample processing
- analysis
1. Bioaerosol characterization
Bioaerosol characterization
Bioaerosol characterization in organic waste treatment facilities is still a controversial topic, since the determination of the biological composition is strongly influenced by environmental factors, such as temperature, humidity, season, and prevalent source contamination, as well as technical factors like sampling and analysis methods. It is not yet possible to define a standardized method for bioaerosol characterization, although some identified procedures that can lead to a more homogeneous research and could allow a more exhaustive bioaerosol description. The critical points of the pipeline characterization are discussed here. Figure 1 reports a summary of the main preliminary steps to conduct a bioaerosol risk assessment.
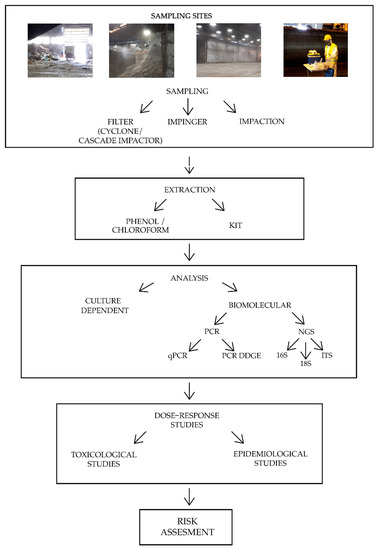
Figure 1. Summary of the main bBioaerosol charisk assessment steps. The four pictures at the top represent common sampling sitesacterization in organic waste treatment plants. Note: PCR = Polymerase Chain Reaction; qPCR = quantitative Polymerase Chain Reaction; PCR DGGE = Polymerase Chain Reaction Denaturing Gradient Gel Electrophoresis; NGS = Next Generation Sequencing; 16 S = ribosomal region 16S of bacteria; 18 S = ribosomal region 18S of fungi; ITS = Internal Transcribed Spacer.
2. Sampling and Extraction Method
Resufacilities is still a controversial ts from exposure studies are extremely influenced by the sampling method. In the last decade, the number of samplers for bioaerosol has increased. Sampling methods commonly used and regulated by international standardized operative procedures are filtration, impaction, liquid impinger, and cyclone[1][2][3].
Filtration consists oopic, since the determination of pumping air through a porous membrane filter that captures bioaerosol particles. This method can be used both in culture-dependent and culture-independent studies. This sampler is cheap, easy to use, efficient at trapping microorganisms larger than a pore size, and the captured microorganisms remain viable[4]. Same biological comples could be subjected to desiccation due to the flow rate and time; therefore, the spore-forming microorganisms may be preferentially recovered[5]. Gesition is stronerally, a personal sampler runs at a low-flow sampling rate, and only a small volume of air can be sampled on the filter. The sample of bioaerosol is not concentrated enough for an exhaustive evaluation; therefore, the enly influenced by environmental sampling can supply another element for risk assessment[6][7].
Impaction consists in the factors, suse of an air pump that captures the air over the surface of a Petri dish containing nutrient agar. Fungal and bacterial samples can be used for culture-dependent studies; it is also easy to use and cheap. The disadvantages are loss of viability and loss of recovery efficiency due to the lack of adherence to the surface by the microorganisms[4].
Liquid impinger ch as temperature, humidity, season, and prevalent source contaminaptures bioaerosol in a liquid matrix. Samples can be analyzed with culture-dependent and -independent methods. These samplers are made of glass, and they can be easily broken, meaning that they are not easy to use, and they are expensive. Samples could be subjected to evaporation and violent bubbling, which could lead to a loss of sample[5].
Cyclone captures bion, as well as technical factors like samplioaerosol into a liquid matrix using centrifugal force. Samples can be analyzed with culture-dependent and -independent techniques. As observed for the impinger method, one of the main disadvantages is theg and analysis methods. It is not yet possible evaporation of the liquid medium and the subsequent loss of sample. Another one is the low quantity of sample collected. Moreover, larger molecules are preferentially collected. This sampler is easy to sterilito define a standardize[7][8].
Nowadays, researchers are trying to improve the real-time bioaerosol monitoring. The currently used devices only count particles but are not able to classify them. The last improvement of such technique included biochemical determination to detect biological components even if there are a lot of non-biological interferents[9].method for bioaerosol characterization, The ideal real-time sampler should be portable, have continuous sampling and high collection efficiency, and should continuously deliver samples to the detection system. The slow development in this field is due to the difficulty in determining the concentration and the type of bioaerosols in the air stream. One of the main disadvantages is the misinterpretation due to the signal interference from non-biological particlethough some identified procedures[10].
The first phase of this study was focused on the comparison of the commonly used sampling methods (Table 1, first section referring to sampling methods), tested both on samples generated in lab and on samples from a composting site. Ferguson et al. observed that Filtration with polycarbonate (PC) filters prodhat can lead to a more homogeneouces better results in terms of bacterial DNA recovery than Liquid impinger with a difference of an order of magnitude[11]. This is probabresearch and couly due to the easier extraction of the sample from the surface of the filter or to the recovery of particles with a diameter size of less than 0.5 µm, which results extremely difficult with the impinger method. Furthermore, filters in PC are thin; hence, they completely dissolve in phenol/chloroform solution making the extraction lysis process more efficient[11]. Despite the higher DNA extraction, in fi allow a more exhaustive bioaerosoltration, the bacteria quantification was scarce and the quality of the DNA was low compared to the Impaction method (Low Melting Agar)[11]description. This could be due to the not optimal extraction methods or to the strong dehydration during high flow sampling rate. It is also important to define an ideal flow rate and sampling time in order to reduce the sample loss due to the desiccation. On the other hand, too short sampling times and low flow sampling rates produce an insufficient collection of bioaerosol for proper extraction and analysis. The use of an optimized extraction kit could make this process easier, time-saving, and would also allow the use of a standardizedcritical points of the pipeline characterization are discussed here. extraction method common for all laboratoriesFigure 1 reports a summary of the main preliminary steps to conduct a bioaerosol risk assessment.

Sampling and Extraction Method
Table 1.
| Methods | Strength | Weakness |
|---|
| Sampling | Filtration | High collection efficiency (for particles >0.4 μm); Portability; Viable microorganisms; Cultural and biomolecular analysis Variable flow rates, it depends on the employed sampler (low 2 L/min; medium 10–30 L/min or high 300–1000 L/min) Also virus collection |
Spore forming microorganism’s selection; Dehydrating problem at high flow rate The low and medium flow produce a scarce quantity of material for the bioaerosol analysis |
||
| Impaction | Variable flow rate but generally <200 L/min; Portability; Generally, only cultural method |
Difficult nucleic acids extraction; Viability reduction; Recovery reduction |
|||
| Impinger | Low flow rates (<100 L/min); Shorter sampling period; Cultural and biomolecular analysis; Sample directly in liquid |
Lower collection efficiency than filters; Low portability; Difficult nucleic acids extraction; Evaporation bias Less cheap |
|||
| Extraction | Phenol-chloroform | Better lysis Cheap |
Not standardized Time-consuming |
||
| Commercial kits (DNeasy PowerSoil Kit Qiagen, PowerViral DNA/RNA Kit Qiagen, etc.) |
Standardized method Time-saving |
Less cheap | |||
| Current Analysis | Cultural | Viable microorganisms; Cheap; Possible API | ® | determination | Qualitative analysis; Do not detect unculturable microorganisms; Detection of 1.5–15.3% of all the species |
| Real time qPCR | Quantitative analysis; High sensitivity and accuracy; High reproducibility; High number of samples; Detect unculturable microorganisms |
Detect also non-viable microorganisms | |||
| PCR-DGGE | Numerically dominant community members; Community changes and differences |
Weak reproducibility; Low specificity; Qualitative analysis; Gradient production |
|||
| MALDI-TOF | Rapid analysis after the growth on plate Sensitive Cheap if the equipment is already available |
Previous cultural step or bioaerosol sample pre-treatment; Identification limited to known peptide mass fingerprints |
|||
| NGS Analysis | Targeted Amplicon Sequencing rRNA | Bacterial and fungal sequencing; Reduction of bias with degenerated oligonucleotides; Most used method |
High sample quantity; Previous PCR step; Expensive; Over-estimation; Detect non-viable microorganisms; Relative abundance sloped; No viral identification |
||
| Shotgun metagenomics | Whole genome Viral detection Taxonomic biodiversity and biological functions Global biome characterization Absolute abundance No PCR biases No previous knowledge of the sequences |
High sample quantity; Expensive; Challenging bioinformatics analyses; Detect non-viable microorganisms |
3. Analytical Methods
Analytical Methods
The commonly used methods for bioaerosol characterization are culture-dependent and quantitative Polymerase Chain Reaction (qPCR) techniques. Moreover, some studies included Next Generation Sequencing (NGS) of the regions 16S and 18S of rRNA or rDNA (Table 1).These three methods do not exclude each other; in fact, the use of all of them allows to increase the comprehension of the bioaerosol composition. The cultural method only recognizes 1.5–15.3% of all the species that are able to create a colony, and all the microorganisms that are not viable or unable to grow are not identified
[29]. In addition, a metagenomic approach could be theoretically applicable; nowadays, there are no published papers where the metagenomic method was applied to bioaerosol composting samples. Such approach could provide a more exhaustive microbiota characterization including also fungi and viruses, but on the other hand, it is still expensive. Traditional culture-dependent studies require selective culture media that allow the growth of target microorganisms limiting the undesired ones, but in the practice, the biodiversity of the matrix highlights overlapping and unexpected growth.
Bacterial and fungal quantifications are a preliminary good contamination index
[29]
Legionella pneumophila, Saccharopolyspora rectivirgula[30]
[32]
[33]
[34]
Polymerase Chain Reaction
[24].
A major disadvantage of qPCR is that it does not determine cell viability. However, a viability assay that combines qPCR with propidium monoazide (PMA-qPCR) can set alive and dead cells apart. Propidium monoazide, as well as ethidium monoazide, is a DNA dye able to bind free DNA avoiding the subsequent PCR reaction
A major disadvantage of qPCR is that it does not determine cell viability. However, a viability assay that combines qPCR with propidium monoazide (PMA-qPCR) can set alive and dead cells apart. Propidium monoazide, as well as ethidium monoazide, is a DNA dye able to bind free DNA avoiding the subsequent PCR reaction
[35]
[36].
The
Polymerase Chain Reaction-Denaturing Gradient Gel Electrophoresis
Next Generation Sequencing (NGS) techniques. However, it allows a primary investigation of the numerically dominant community members, community changes, and differences. It has a scarce reproducibility and sensitivity though.
NGS is one of the latest technological innovations introduced in the biomolecular practice. The main techniques used are 454 pyrosequencing and Illumina.
NGS is one of the latest technological innovations introduced in the biomolecular practice. The main techniques used are 454 pyrosequencing and Illumina.
Pyrosequencing is based on an emulsion PCR on microbeads. Each bead covered with DNA amplicons is contained in a plot slightly larger than the beads. On the bottom of the support there is a light detector
[37]
[23]. Moreover, it is also possible to conduct whole genome sequencing, allowing a global analysis of the genome sample. It requires high sample quantities, it is expensive, and the bioinformatic analyses are still challenging. On the other hand, it allows a global microbiome characterization (e.g., virome), including taxonomic biodiversity and biological functions information.
Few metagenomic studies on bioaerosol were published. The collection of adequate biomass is crucial for successful metagenomic analysis. Recently, metagenomic analysis of the airborne DNA and RNA were performed in a daycare center, starting from the filters of the ventilation and the air conditioning system
[40]
the different methods used[41].
[30].
The cultural method, especially if preceded by an enrichment phase, is able to identify microorganisms that are present in the environment at very low concentrations, thus allowing the tracing of potentially pathogenic microorganisms. A culture-dependent confirmation and typing with culture-independent techniques are necessary to establish a causal link between the infectious episode and the exposure. The analysis method is truly complete only when both the biomolecular and cultural studies are combined
[42].
Matrix-assisted laser desorption/ionization time of flight (MALDI-TOF)
[18]
[22]
[21]
[16]
[17].
4. Risk Assessment
Risk Assessment
To perform risk assessments, some scientists decided to investigate the presence of harmful microorganisms for human health using qPCR. Dubuis et al. selected four bacterial biomarkers that have been shown in composting facilities in order to verify their presence also in biomethanization areas:Saccharopolyspora rectivirgula, Legionella
spp.,Legionella pneumophila
, andMycobacterium
spp.[30]
.S. rectivirgula
is correlated with the onset of the Farmer’s lung disease;L. pneumophila
causes a serious disease, the Legionnaires’ disease[44]
;Mycobacterium
spp. can cause bone, skin, and lung infections.Mbareche et al. focused their attention on the study of fungal composition of bioaerosols emitted from biomethanization sites and decided to use
Aspergillus
Aspergillus fumigatus as bioindicators .
The investigation of
A. fumigatus
[45].
Moreover, Traversi et al. used specific bioindicators such as Pseudomonaceae to improve the risk assessment in biomethanization plants
[6]
Pseudomonas
[6].
Nowadays, an exhaustive biological risk assessment is weakly applied because it is strongly influenced by parameters that are difficult to estimate and that are often arbitrarily defined, such as the variability of microorganisms present in the bioaerosol, the rate of bioaerosol released from the matrix, the inter-individual variability of the exposed subjects, etc. In addition, the lack of validated methodology causes a deformity in the evaluation that makes the results incomparable.
Nowadays, an exhaustive biological risk assessment is weakly applied because it is strongly influenced by parameters that are difficult to estimate and that are often arbitrarily defined, such as the variability of microorganisms present in the bioaerosol, the rate of bioaerosol released from the matrix, the inter-individual variability of the exposed subjects, etc. In addition, the lack of validated methodology causes a deformity in the evaluation that makes the results incomparable.
Mbareche et al. suggested the creation of a worldwide database containing all the sequences obtained from the sequencing analysis with the description of the sampling and analysis methods used. This would allow a definition of the microbial composition of bioaerosol
[48].
5. Limitations
Limitations
The sampling method based on filters is currently the most satisfying, but it requires improvement and standardization of the extraction methods. The biomolecular analysis allows a better bioaerosol characterization, but this technique does not distinguish between viable or non-viable microorganisms, potentially leading to an overestimation. Another important issue for qPCR application is that microorganisms could have more than one copy of the region 16S, as previously demonstrated in the literature[30]. Therefore, the data elaboration has to take into account such evidence whenever there are quantitative purposes.
. Therefore, the data elaboration has to take into account such evidence whenever there are quantitative purposes.References
- EN 13098. Guidelines for Measurement of Airborne Microorganisms and Endotoxin. Available online:https://shop.bsigroup.com/ProductDetail/?pid=000000000030037644 (accessed on 28 April 2020).
- CEN/TS 16115-1. Measurement of Bioaerosols. Part. 1: Determination of Moulds Using Filter SamplingSystems and Culture-Based Analyses. Available online: https://www.sis.se/en/produkter/environmenthealth-protection-safety/air-quality/ambient-atmospheres/siscents1611512011/ (accessed on 28 April 2020).
- EN 1403. Determination of Airborne Endotoxins. Available online: http://store.uni.com/catalogo/en-14031-2003?josso_back_to=http://store.uni.com/josso-security-check.php&josso_cmd=login_optional&josso_partnerapp_host=store.uni.com/ (accessed on 28 April 2020).
- Dale W. Griffin; Atmospheric Movement of Microorganisms in Clouds of Desert Dust and Implications for Human Health. Clinical Microbiology Reviews 2007, 20, 459-477, 10.1128/cmr.00039-06.
- Keunje Yoo; Tae Kwon Lee; Eun Joo Choi; Jihoon Yang; Sudheer Kumar Shukla; Sang-Il Hwang; Joonhong Park; Molecular approaches for the detection and monitoring of microbial communities in bioaerosols: A review. Journal of Environmental Sciences 2017, 51, 234-247, 10.1016/j.jes.2016.07.002.
- Deborah Traversi; Ilaria Gorrasi; Claudio Pignata; Raffaella Degan; Elisa Anedda; Giulia Carletto; Greta Vercellino; Stefania Fornasero; Antonino Bertino; Francesca Filippi; et al.Maria GulloGiorgio Gilli Aerosol exposure and risk assessment for green jobs involved in biomethanization. Environment International 2018, 114, 202-211, 10.1016/j.envint.2018.02.046.
- Bipasha Ghosh; Himanshu Lal; Arun Srivastava; Review of bioaerosols in indoor environment with special reference to sampling, analysis and control mechanisms.. Environment International 2015, 85, 254-72, 10.1016/j.envint.2015.09.018.
- Haig, C.W.; Mackay,W.G.; Walker, J.T.; Williams, C.; Bioaerosol sampling: Sampling mechanisms, bioeffciency and field studies.. J. Hosp. Infect. 2016, 93, 242-255.
- Tian, J.H.; Yan, C.; Nasir, Z.A.; Alcega, S.G.; Tyrrel, S.; Coulon, F.; Real time detection and characterisation of bioaerosol emissions from wastewater treatment plants. Sci. Total Environ. 2020, 721, 137629.
- Yu Sung Cho; Seung Chan Hong; Jeongan Choi; Jae Hee Jung; Development of an automated wet-cyclone system for rapid, continuous and enriched bioaerosol sampling and its application to real-time detection. Sensors and Actuators B: Chemical 2019, 284, 525-533, 10.1016/j.snb.2018.12.155.
- Robert Ferguson; Sonia Garcia Alcega; Frédéric Coulon; Alex J. Dumbrell; Corinne Whitby; Ian Colbeck; Bioaerosol biomonitoring: Sampling optimization for molecular microbial ecology. Molecular Ecology Resources 2019, 19, 672-690, 10.1111/1755-0998.13002.
- Jonathan Pilote; Valérie Létourneau; Matthieu Girard; Caroline Duchaine; Quantification of airborne dust, endotoxins, human pathogens and antibiotic and metal resistance genes in Eastern Canadian swine confinement buildings. Aerobiologia 2019, 35, 283-296, 10.1007/s10453-019-09562-6.
- Mbareche, H.; Veillette, M.; Bilodeau, G.J.; Duchaine, C.; Bioaerosol sampler choice should consider effciency and ability of samplers to cover microbial diversity.. Appl. Environ. Microbiol. 2018, 84, 1-22.
- Wenjun Jiang; Peng Liang; Buying Wang; Jianhuo Fang; Jidong Lang; Geng Tian; Jingkun Jiang; Ting F. Zhu; Optimized DNA extraction and metagenomic sequencing of airborne microbial communities. Nature Protocols 2015, 10, 768-779, 10.1038/nprot.2015.046.
- Philippe Duquenne; On the Identification of Culturable Microorganisms for the Assessment of Biodiversity in Bioaerosols. Annals of Work Exposures and Health 2017, 62, 139-146, 10.1093/annweh/wxx096.
- Anne Mette Madsen; Margit W. Frederiksen; Mikkel Hyldeqvist Jacobsen; Kira Tendal; Towards a risk evaluation of workers’ exposure to handborne and airborne microbial species as exemplified with waste collection workers. Environmental Research 2020, 183, 109177, 10.1016/j.envres.2020.109177.
- Zaheer Ahmad Nasir; Catherine Rolph; Samuel Collins; David Stevenson; Toni Gladding; Enda Hayes; Ben Williams; Shagun Khera; Simon K. Jackson; Allan Bennett; et al.Simon ParksR.P KinnersleyKerry WalshSimon J. T. PollardGillian DrewSonia Garcia AlcegaFrédéric CoulonSean Tyrrel A Controlled Study on the Characterisation of Bioaerosols Emissions from Compost. Atmosphere 2018, 9, 379, 10.3390/atmos9100379.
- Neelja Singhal; Manish Kumar; Pawan Kumar Kanaujia; Jugsharan Singh Virdi; MALDI-TOF mass spectrometry: an emerging technology for microbial identification and diagnosis. Frontiers in Microbiology 2015, 6, 1-16, 10.3389/fmicb.2015.00791.
- Anne Mette Madsen; Athanasios Zervas; Kira Tendal; Jeppe Lund Nielsen; Microbial diversity in bioaerosol samples causing ODTS compared to reference bioaerosol samples as measured using Illumina sequencing and MALDI-TOF. Environmental Research 2015, 140, 255-267, 10.1016/j.envres.2015.03.027.
- Jennie Cox; Hamza Mbareche; William G. Lindsley; Caroline Duchaine; Field sampling of indoor bioaerosols. Aerosol Science and Technology 2019, 54, 572-584, 10.1080/02786826.2019.1688759.
- Anne Mette Madsen; Trine Thilsing; Jesper Bælum; Anne Helene Garde; Ulla Vogel; Occupational exposure levels of bioaerosol components are associated with serum levels of the acute phase protein Serum Amyloid A in greenhouse workers.. Environmental Health 2016, 15, 9, 10.1186/s12940-016-0090-7.
- Katharina Druckenmüller; Andrea Gärtner; Udo Jäckel; Kerstin Klug; Johannes Schiffels; Klaus Günther; Gereon Elbers; Development of a methodological approach for the characterization of bioaerosols in exhaust air from pig fattening farms with MALDI-TOF mass spectrometry. International Journal of Hygiene and Environmental Health 2017, 220, 974-983, 10.1016/j.ijheh.2017.05.003.
- Núñez, A.; de Paz, G.A.; Rastrojo, A.; García, A.M.; Alcamí, A.; Montserrat Gutiérrez-Bustillo, A.; Moreno, D.A.; Monitoring of airborne biological particles in outdoor atmosphere. Part 2: Metagenomics applied to urban environments. Int. Microbiol. 2016, 19, 69-80.
- Heid, C.A.; Stevens, J.; Livak, K.J.; Williams, P.M.; Real time quantitative PCR. Genome Res. 1996, 6, 986-994.
- Hamza Mbareche; Evelyne Brisebois; Marc Veillette; Caroline Duchaine; Bioaerosol sampling and detection methods based on molecular approaches: No pain no gain. Science of The Total Environment 2017, 599, 2095-2104, 10.1016/j.scitotenv.2017.05.076.
- Shendure, J.; Ji, H.; Next-generation DNA sequencing. Nat. Biotechnol. 2088, 26, 1135-1145.
- Rob Knight; Alison Vrbanac; Bryn C. Taylor; Alexander Aksenov; Chris Callewaert; Justine Debelius; Antonio González; Tomasz Kosciolek; Laura-Isobel McCall; Daniel McDonald; et al.Alexey V. MelnikJames T. MortonJosé NavasRobert A. QuinnJon G. SandersAustin D. SwaffordLuke R. ThompsonAnupriya TripathiZhenjiang Z. XuJesse R. ZaneveldQiyun ZhuJ. Gregory CaporasoPieter C. Dorrestein Best practices for analysing microbiomes. Nature Reviews Microbiology 2018, 16, 410-422, 10.1038/s41579-018-0029-9.
- Vigdis Torsvik; Frida Lise Daae; Ruth-Anne Sandaa; L Ovreås; Novel techniques for analysing microbial diversity in natural and perturbed environments. Journal of Biotechnology 1998, 64, 53-62, 10.1016/s0168-1656(98)00103-5.
- Nathalie Wéry; Bioaerosols from composting facilities—a review. Frontiers in Microbiology 2014, 4, 1-9, 10.3389/fcimb.2014.00042.
- Marie-Eve Dubuis; Hamza Mbareche; Marc Veillette; Bouchra Bakhiyi; J. Zayed; J. Lavoie; Caroline Duchaine; Bioaerosols concentrations in working areas in biomethanization facilities. Journal of the Air & Waste Management Association 2017, 67, 1258-1271, 10.1080/10962247.2017.1356762.
- Ming-Wei Chang; Chung-Ru Lee; Hsueh-Fen Hung; Kuo-Sheng Teng; Hsin Huang; Chun-Yu Chuang; Bioaerosols from a Food Waste Composting Plant Affect Human Airway Epithelial Cell Remodeling Genes. International Journal of Environmental Research and Public Health 2013, 11, 337-354, 10.3390/ijerph110100337.
- Laetitia Betelli; P. Duquenne; Frédéric Grenouillet; Xavier Simon; Emeline Scherer; Evelyne Gehin; Alain Hartmann; Development and evaluation of a method for the quantification of airborne Thermoactinomyces vulgaris by real-time PCR. Journal of Microbiological Methods 2013, 92, 25-32, 10.1016/j.mimet.2012.10.009.
- Elisa Anedda; Giulia Carletto; Giorgio Gilli; Deborah Traversi; Monitoring of Air Microbial Contaminations in Different Bioenergy Facilities Using Cultural and Biomolecular Methods.. International Journal of Environmental Research and Public Health 2019, 16, 2546, 10.3390/ijerph16142546.
- Ashley Shade; Clifford S. Hogan; Amy K. Klimowicz; Matthew Linske; Patricia S. McManus; Jo Handelsman; Culturing captures members of the soil rare biosphere. Environmental Microbiology 2012, 14, 2247-2252, 10.1111/j.1462-2920.2012.02817.x.
- Joanne B. Emerson; Rachel Adams; Clarisse M. Betancourt Román; Brandon Brooks; David A. Coil; Katherine Dahlhausen; Holly H. Ganz; Erica M. Hartmann; Tiffany Hsu; Nicholas B. Justice; et al.Ivan G. Paulino-LimaJulia C. LuongoDespoina LymperopoulouCinta Gomez-SilvanBrooke Rothschild-MancinelliMelike BalkCurtis HuttenhowerAndreas NockerParag VaishampayanL.J. Rothschild Schrödinger’s microbes: Tools for distinguishing the living from the dead in microbial ecosystems. Microbiome 2017, 5, 86, 10.1186/s40168-017-0285-3.
- Ching-Wen Chang; Nien-Tzu Hung; Nai-Tzu Chen; Optimization and application of propidium monoazide-quantitative PCR method for viable bacterial bioaerosols. Journal of Aerosol Science 2017, 104, 90-99, 10.1016/j.jaerosci.2016.11.002.
- Zhentong Li; H.W. Lu; Lixia Ren; Li He; Experimental and modeling approaches for food waste composting: A review. Chemosphere 2013, 93, 1247-1257, 10.1016/j.chemosphere.2013.06.064.
- Martin Kircher; Janet Kelso; High-throughput DNA sequencing - concepts and limitations. BioEssays 2010, 32, 524-536, 10.1002/bies.200900181.
- Aaron J. Prussin; Linsey C. Marr; Kyle Bibby; Challenges of studying viral aerosol metagenomics and communities in comparison with bacterial and fungal aerosols. FEMS Microbiology Letters 2014, 357, 1-9, 10.1111/1574-6968.12487.
- Aaron J. Prussin; Pedro J. Torres; John Shimashita; Steven Robert Head; Kyle Bibby; Scott T. Kelley; Linsey C. Marr; Seasonal dynamics of DNA and RNA viral bioaerosol communities in a daycare center. Microbiome 2019, 7, 53, 10.1186/s40168-019-0672-z.
- Karren Prost; Harold Kloeze; Shamir Mukhi; Katie Bozek; Zvonimir Poljak; Samira Mubareka; Bioaerosol and surface sampling for the surveillance of influenza A virus in swine. Transboundary and Emerging Diseases 2019, 66, 1210-1217, 10.1111/tbed.13139.
- Alejandra Cerda; Adriana Artola; Xavier Font; Raquel Barrena; Teresa Gea; Antoni Sánchez; Composting of food wastes: Status and challenges. Bioresource Technology 2018, 248, 57-67, 10.1016/j.biortech.2017.06.133.
- Olivier Le Goff; Jean-Jacques Godon; Kim Milferstedt; Hélène Bacheley; J.P. Steyer; Nathalie Wéry; A new combination of microbial indicators for monitoring composting bioaerosols. Atmospheric Environment 2012, 61, 428-433, 10.1016/j.atmosenv.2012.07.081.
- S. Casati; L. Conza; J. Bruin; V. Gaia; Compost facilities as a reservoir of Legionella pneumophila and other Legionella species. Clinical Microbiology and Infection 2010, 16, 945-947, 10.1111/j.1469-0691.2009.03009.x.
- Hamza Mbareche; Marc Veillette; Marie-Eve Dubuis; Bouchra Bakhiyi; Geneviève Marchand; J. Zayed; J. Lavoie; Guillaume J. Bilodeau; Caroline Duchaine; Fungal bioaerosols in biomethanization facilities. Journal of the Air & Waste Management Association 2018, 68, 1198-1210, 10.1080/10962247.2018.1492472.
- Alba Ribera; Eva Benavent; Jaime Lora-Tamayo; Fe Tubau; Salvador Pedrero; Xavier Cabo; J. Ariza; Oscar Murillo; Osteoarticular infection caused by MDRPseudomonas aeruginosa: the benefits of combination therapy with colistin plus β-lactams. Journal of Antimicrobial Chemotherapy 2015, 70, 3357–3365, 10.1093/jac/dkv281.
- So-Youn Park; Hyun Jung Park; Song Mi Moon; Ki-Ho Park; Yong Pil Chong; Mi-Na Kim; Sung-Han Kim; Sang-Oh Lee; In-Hwan Oh; Jun Hee Woo; et al.Sang-Ho Choi Impact of adequate empirical combination therapy on mortality from bacteremic Pseudomonas aeruginosa pneumonia. BMC Infectious Diseases 2012, 12, 308-308, 10.1186/1471-2334-12-308.
- Hamza Mbareche; Lidia Morawska; Caroline Duchaine; On the interpretation of bioaerosol exposure measurements and impacts on health. Journal of the Air & Waste Management Association 2019, 69, 789-804, 10.1080/10962247.2019.1587552.
