To protect the cell from the accumulation of aberrant proteins, a network of protein quality control (PQC) pathways identifies the substrates and direct them towards refolding or elimination via regulated protein degradation. The main pathway for degradation of misfolded proteins is the ubiquitin-proteasome system. PQC pathways have been first described in the cytoplasm and the endoplasmic reticulum, however, accumulating evidence indicates that the nucleus is an important PQC compartment for ubiquitination and proteasomal degradation of not only nuclear, but also cytoplasmic proteins.
- proteasome
- ubiquitin
- nucleus
- inner nuclear membrane
- protein misfolding
Thanks so much for your check. We sincerely hope you may create this entry based on your published paper. You can click the “submit” button to upload it and revise it. We will help you layout after you submit it. Moreover, we will link your article at the entry, and more scholars and students can look through it.
1. Introduction
Maintaining a functional proteome, or proteostasis, is one of the key tasks in the cell. Proteins that are aberrant, for instance due to misfolding, inability to form complexes, or incorrect localization, can be harmful for the cell as a result of a loss of function, interference with other processes or inappropriate interactions with other components in the cell [1][2][1,2]. Proteins are at risk of misfolding especially during protein synthesis and assembly into higher-order structures or protein complexes [3]. The effect of protein misfolding and aggregation is particularly evident in proteinopathies, including neurodegenerative diseases such as Alzheimer’s, Parkinson’s, Huntington’s, amyotrophic lateral sclerosis, and others, where accumulation of protein aggregates is a hallmark of pathology. Cells have developed an intricate network of protein quality control (PQC) pathways by which they facilitate folding, assess protein quality, and in the case of an aberrant protein, initiate a proper response to mitigate the damage, either by refolding, or by eliminating the protein via degradation pathways. The main pathway for degradation of misfolded proteins is the ubiquitin-proteasome system.
Degradation-mediated PQC mechanisms have been best described in the cytoplasm and endoplasmic reticulum (ER) [4], however nucleus has emerged as a key PQC compartment for ubiquitination and proteasomal degradation of not only nuclear, but also cytoplasmic proteins [5]. In cells, proteasomes are localized in the cytoplasm, as well as in the nucleus [6][7][8][9][6,7,8,9]. In fact, in proliferating yeast cells, the majority of cellular proteasomes are localized in the nucleus [10]. Nucleus and the inner nuclear membrane (INM) contain ubiquitination machinery involved in PQC pathways that are important for proteostasis maintenance. In this paper, we summarize recent findings on nuclear ubiquitin-proteasome pathways that function in PQC in Saccharomyces cerevisiae: The INM-associated degradation (INMAD) mediated by the E3 ubiquitin ligases Asi1-3, Doa10, and APC/C, and a nuclear pathway for degradation of misfolded proteins mediated by the E3 ubiquitin ligase San1 (Figure 1 and Table 1).
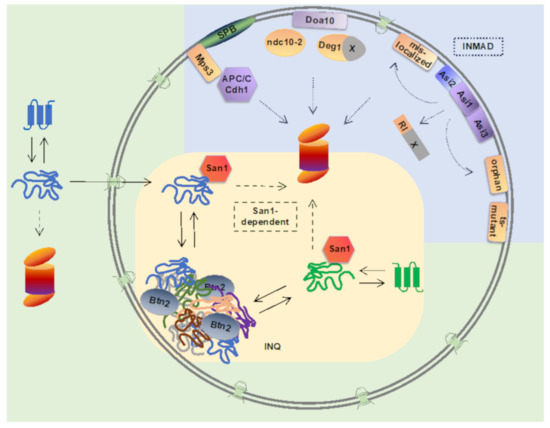
Figure 1. Nuclear ubiquitin-proteasome-dependent protein quality control pathways in yeast. Inner nuclear membrane-associated degradation (INMAD) is mediated via E3 ubiquitin ligases Asi1-3, Doa10, and APC/C. Integral INM-localized Asi-complex consists of E3 ubiquitin ligases Asi1 and Asi3, and a substrate-specific adaptor protein Asi2. Asi-complex targets nuclear RI-degron-containing proteins. Integral membrane Asi-substrates include proteins mislocalized to the INM, orphan subunits of unassembled proteins complexes, and temperature-sensitive (ts) mutants. Integral membrane E3 ligase Doa10 localizes to both the endoplasmic reticulum and the INM, and its substrates include Deg1-degron containing proteins, Ndc10-2 kinetochore mutant protein, and INM protein Asi2. E3 ligase APC/C with its co-activator Cdh1 target Mps3, an integral INM protein of the spindle pole body (SPB). Nuclear E3 ligase San1 targets misfolded cytoplasmic and nuclear proteins for proteasomal degradation. Upon ubiquitin proteasome system overload, misfolded nuclear and cytoplasmic proteins can be reversibly sequestered into intranuclear quality compartment (INQ) by the sequestrase Btn2. Upon disaggregation, misfolded proteins can be directed to refolding or degradation.
2. Ubiquitin-Proteasome System
The main site for degradation of misfolded and short-lived proteins is the proteasome, a multiprotein proteolytic machine consisting of a 20S catalytic core particle, and one or two 19S regulatory particles that recognize proteins marked for destruction and regulate substrate entry into the core [11]. Proteins are tagged for proteasomal degradation by attachment of ubiquitin, a small, highly conserved globular protein, which is recognized by ubiquitin binding proteins in the regulatory particle of the proteasome. Regulatory particle additionally contains ATPases that unfold the substrate and translocate it into the 20S chamber for proteolysis by three distinct enzymatic activities, resulting in substrate cleavage into short peptides. The core tunnel is narrow, and as a consequence, proteins must be unfolded prior to degradation by the proteasome [12]. Due to the physical constraints of the 20S core tunnel, the proteasome is able to degrade individual proteins, while protein aggregates and larger structures can be degraded by autophagy [13]. The role of autophagy in degradation of nuclear components has been reviewed elsewhere [14].
Ubiquitin is covalently attached to the protein substrate in a series of enzymatic reactions catalyzed by ubiquitin activating enzyme (E1), ubiquitin-conjugating enzyme (E2), and E3 ubiquitin-protein ligase, usually to the lysine residue side chains [15]. Proteins that have failed to attain a proper structure due to genetic mutations, errors in translation or environmental stress, display degradation signals, or degrons [16], protein segments that are often characterized by exposed stretches of hydrophobic residues. Normally, hydrophobic peptides are buried within the folded protein, localized at the interface with interacting proteins, or embedded within a membrane layer, but may become exposed due to protein misfolding, truncation, or a lack of interaction partner. These signals are recognized by the ubiquitination machinery, often assisted by the molecular chaperones, and the main determinants of protein substrate selectivity are E3 ubiquitin protein ligases [17].
Prior to the delivery to the proteasome, many polyubiquitinated proteins present in protein complexes or embedded within membranes need to be first extracted by the Cdc48 ATPase complex [18]. Cdc48 is a conserved ATP-ase of the AAA+ family (ATPases associated with a variety of cellular activities) whose cellular functions are determined by its association with many different cofactors, including a heterodimer formed by Ufd1 and Npl4 [19]. Within the Cdc48-Ufd1-Npl4 complex, co-factor Npl4 is responsible for recognizing polyubiquitinated substrates, with a strong preference for the K48-type ubiquitin chains [20]. Cdc48-complex bound polyubiquitinated substrate is extracted by passing through the central pore of the Cdc48 homo-hexameric ring, by the ATP hydrolysis, and is subsequently released from the Cdc48-complex.
Polyubiquitinated substrates may be bound by the proteasome directly, by the ubiquitin receptors present in the proteasome regulatory particle. Alternatively, polyubiquitinated substrates could be delivered to the proteasome indirectly, via ubiquitin-like (UbL) and ubiquitin-associated (UBA) family of ubiquitin binding proteins, represented by Dsk2, Rad23, and Ddi1 in yeast [21]. The UBA domain of these proteins binds to ubiquitin on the modified substrates, while their UbL domains bind to ubiquitin receptors at the proteasome regulatory particle [21]. UbL-UBA proteins thus serve as adaptors that link ubiquitinated substrate proteins to the proteasome, delivering them for degradation [21]. The data from a recent study indicate that a large proportion of ubiquitinated proteasome substrates are delivered to the proteasomes indirectly, by UbL-UBA proteins Rad23 and Dsk2 [20].
Substrate-bound polyubiquitin chains can be trimmed or removed by the activity of deubiquitinating enzymes (DUBs), which hydrolyze the bond between substrate and the ubiquitin, or between two ubiquitin molecules [22]. Yeast genome encodes around 20 different DUBs that are localized in ER, mitochondria, nucleus, and cytoplasm, including two DUBs, Ubp6 and Rpn11, that are associated with the proteasome regulatory particle [23]. Different functions of DUBs include protein stabilization or reversal of ubiquitin signaling by the removal of ubiquitin chains from target proteins, editing the ubiquitin modification by trimming the polyubiquitin chains and ubiquitin recycling [22]. A recent screen examining the role of DUBs in protein quality control showed that the degradation of cytosolic quality control substrates is affected by a wide range of the DUB deletion mutants, furthermore the ER-associated degradation is affected by deletion of a DUB gene UBP3, together indicating the involvement of DUBs in the quality control pathways [24].
The ubiquitin-proteasome pathways, including the proteasome, ubiquitination, and deubiquitination machinery, chaperones, and accessory proteins, are conserved from yeast to human [17].
3. The Nucleus and the Nuclear Envelope at a Glance
The nucleus is enclosed by the nuclear envelope (NE), which consists of two lipid bilayers, the inner and the outer nuclear membrane (INM and ONM) [25]. INM and ONM are joined together at the sites of nuclear pore complexes, which allow nucleocytoplasmic transport. While the ONM is continuous with the ER membrane, the INM has a specific protein composition that considerably differs from that of the ONM and the ER. In metazoan cells the nuclear side of the INM is lined by a meshwork of lamin intermediate filaments and lamin-interacting proteins, called the nuclear lamina [26][27][26,27].
Following their co- or post-translational insertion into the ER membrane, integral membrane proteins destined to the INM are able to diffuse from the ER membrane to the ONM and can gain access to the INM via the nuclear pore membrane [28]. Although many mechanistic details of INM protein transport are still unclear, most INM proteins appear to reach the INM by diffusion, with their extraluminal domains passing through the central or lateral channels of the nuclear pore complex and can be retained in the nucleus via interaction with nuclear components, such as lamins or chromatin [28]. INM targeting of certain integral membrane proteins requires active transport, similar to the pathway used by soluble cargo [29].
Reference (we'll rearrange the references after you submitted it)
- Chiti, F.; Dobson, C.M. Protein Misfolding, Amyloid Formation, and Human Disease: A Summary of Progress Over the Last Decade. Annu. Rev. Biochem. 2017, 86.
- Valastyan, J.S.; Lindquist, S. Mechanisms of protein-folding diseases at a glance. Dis. Model. Mech. 2014, 7, 9–14.
- Collins, G.A.; Goldberg, A.L. The Logic of the 26S Proteasome. Cell 2017, 169, 792806.
- Buchberger, A.; Bukau, B.; Sommer, T. Protein Quality Control in the Cytosol and the Endoplasmic Reticulum: Brothers in Arms. Mol. Cell 2010, 40, 238–252.
- Nielsen, S.V.; Poulsen, E.G.; Rebula, C.A.; Hartmann-Petersen, R. Protein quality control in the nucleus. Biomolecules 2014, 4, 646–661.
- Lafarga, M.; Fernández, R.; Mayo, I.; Berciano, M.T.; Castaño, J.G. Proteasome dynamics during cell cycle in rat Schwann cells. Glia 2002, 38, 313–328.
- Rivett, A.J.; Palmer, A.; Knecht, E. Electron Microscopic Localization of the Multicatalytic Proteinase Complex in Rat Liver and in Cultured Cells. J. Histochem. Cytochem. 1992, 40, 1165–1172.
- Reits, E.A.J.; Benham, A.M.; Plougastel, B.; Neefjes, J.; Trowsdale, J. Dynamics of proteasome distribution in living cells. EMBO J. 1997, 16, 6087–6094.
- Ádori, C.; Low, P.; Moszkovkin, G.; Bagdy, G.; László, L.; Kovács, G.G. Subcellular distribution of components of the ubiquitin-proteasome system in non-diseased human and rat brain. J. Histochem. Cytochem. 2006, 54, 263–267.
- Russell, S.J.; Steger, K.A.; Johnston, S.A. Subcellular localization, stoichiometry, and protein levels of 26 S proteasome subunits in yeast. J. Biol. Chem. 1999, 274, 21943–21952.
- Ciechanover, A. Intracellular protein degradation: From a vague idea thru the lysosome and the ubiquitin-proteasome system and onto human diseases and drug targeting. Best Pr. Res. Clin. Haematol. 2017, 30, 341–355.
- Finley, D.; Chen, X.; Walters, K.J. Gates, Channels, and Switches: Elements of the Proteasome Machine. Trends Biochem. Sci. 2016, 41, 77–93.
- Yin, Z.; Pascual, C.; Klionsky, D.J. Autophagy: Machinery and regulation. Microb. Cell 2016, 3, 588–596.
- Boban, M.; Foisner, R. Degradation-mediated protein quality control at the inner nuclear membrane. Nucleus 2016, 7, 41–49.
- Ciechanover, A. Proteolysis: From the lysosome to ubiquitin and the proteasome. Nat. Rev. Mol. Cell Biol. 2005, 6, 79–86.
- Varshavsky, A. Letter to the Editor Naming a Targeting Signal. Cell 1991, 64, 13–15.
- Glickman, M.H.; Ciechanover, A. The Ubiquitin-Proteasome Proteolytic Pathway: Destruction for the Sake of Construction. Physiol. Rev. 2002, 82, 373–428.
- Rapoport, T.; Bodnar, N. Toward an understanding of the Cdc48/p97 ATPase. F1000Research 2017, 6, 1–10.
- Hänzelmann, P.; Schindelin, H. The interplay of cofactor interactions and post-translational modifications in the regulation of the AAA+ ATPase p97. Front. Mol. Biosci. 2017, 4, 21.
- Tsuchiya, H.; Ohtake, F.; Arai, N.; Kaiho, A.; Yasuda, S.; Tanaka, K.; Saeki, Y. In Vivo Ubiquitin Linkage-type Analysis Reveals that the Cdc48-Rad23/Dsk2 Axis Contributes to K48-Linked Chain Specificity of the Proteasome. Mol. Cell 2017, 66, 488–502.e7.
- Su, V.; Lau, A.F. Ubiquitin-like and ubiquitin-associated domain proteins: Significance in proteasomal degradation. Cell. Mol. Life Sci. 2009, 66, 2819–2833.
- Komander, D.; Clague, M.J.; Urbé, S. Breaking the chains: Structure and function of the deubiquitinases. Nat. Rev. Mol. Cell Biol. 2009, 10, 550–563.
- Suresh, H.G.; Pascoe, N.; Andrews, B. The structure and function of deubiquitinases: Lessons from budding yeast. Open Biol. 2020, 10, 200279.
- Wu, H.; Ng, D.T.W.; Cheong, I.; Matsudaira, P. The degradation-promoting roles of deubiquitinases Ubp6 and Ubp3 in cytosolic and ER protein quality control. PLoS ONE 2020, 15, e0232755.
- Wilson, K.L.; Berk, J.M. The nuclear envelope at a glance. J. Cell Sci. 2010, 123, 1973–1978.
- Schirmer, E.C.; Foisner, R. Proteins that associate with lamins: Many faces, many functions. Exp. Cell Res. 2007, 313, 2167–2179.
- Gruenbaum, Y.; Foisner, R. Lamins: Nuclear intermediate filament proteins with fundamental functions in nuclear mechanics and genome regulation. Annu. Rev. Biochem. 2015, 84, 131–164.
- Katta, S.S.; Smoyer, C.J.; Jaspersen, S.L. Destination: Inner nuclear membrane. Trends Cell Biol. 2014, 24, 221–229.
- King, M.C.; Lusk, C.P.; Blobel, G. Karyopherin-mediated import of integral inner nuclear membrane proteins. Nature 2006, 442, 1003–1007.
